
@charliebell.bsky.social
Senior post-doc in the Faulkner lab
ARC DECRA Fellow
Interested in how gene regulatory mechanisms contribute to biological complexity
ARC DECRA Fellow
Interested in how gene regulatory mechanisms contribute to biological complexity
Pinned

Chromatin-based memory as a self-stabilizing influence on cell identity - Genome Biology
Cell types are traditionally thought to be specified and stabilized by gene regulatory networks. Here, we explore how chromatin memory contributes to the specification and stabilization of cell states...
doi.org
"Everything in epigenetics is circular". Here, we explore how memory that is stored locally on chromatin has a crucial role in stabilising cellular state, updating our view of the epigenetic landscape from one shaped by the genotype, to one moulded by experience
doi.org/10.1186/s130...
doi.org/10.1186/s130...
Reposted
18/ At large genomic distance, long encounters (=the important ones) are only due to loop extrusion; but at short genomic distance they can also be due to random collisions. This is why enhancers are no sensitive to loss of extrusion when they are close, but sensitive when they are far!
September 24, 2025 at 9:45 PM
18/ At large genomic distance, long encounters (=the important ones) are only due to loop extrusion; but at short genomic distance they can also be due to random collisions. This is why enhancers are no sensitive to loss of extrusion when they are close, but sensitive when they are far!
Reposted
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
September 24, 2025 at 9:45 PM
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
Reposted
We are looking for a student to continue our work on chromatin evolution:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...

July 15, 2025 at 11:30 AM
We are looking for a student to continue our work on chromatin evolution:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...
The project with @seanamontgomery.bsky.social will focus on chromatin state readers across eukaryotes.
More info: recruitment.crg.eu/content/jobs...
Reposted
New preprint from the lab 🎉 on the evolution of gene regulatory network complexity, where we tested whether sexual recombination promotes more complex GRNs using biochemically-inspired evolutionary simulations 🧐. Bluetorial from 1st author @chapelmadison.bsky.social 👇
1/ Why are eukaryotic gene regulatory networks (GRNs) so much more complex than prokaryotic ones?
In our new preprint, we investigated the role that recombination – a key step in eukaryotic reproductive strategies – plays in this problem! www.biorxiv.org/content/10.1...
In our new preprint, we investigated the role that recombination – a key step in eukaryotic reproductive strategies – plays in this problem! www.biorxiv.org/content/10.1...

Evolutionary simulations reveal role for genomic recombination in the evolution of gene regulatory network complexity and robustness
The gene regulatory networks (GRNs) of eukaryotes are dramatically more complex than the GRNs of prokaryotes, but we lack a complete picture of the selective pressures that have shaped this difference...
www.biorxiv.org
September 4, 2025 at 12:37 AM
New preprint from the lab 🎉 on the evolution of gene regulatory network complexity, where we tested whether sexual recombination promotes more complex GRNs using biochemically-inspired evolutionary simulations 🧐. Bluetorial from 1st author @chapelmadison.bsky.social 👇
Reposted
Online Now: Regulation of somatic hypermutation by higher-order chromatin structure Online now:
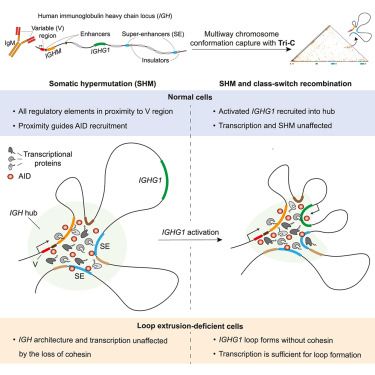
Regulation of somatic hypermutation by higher-order chromatin structure
Schoeberl et al. employ Tri-C to unravel the higher-order chromatin architecture underpinning the co-transcriptional mutagenesis of immunoglobulin genes during antibody maturation in human B cells. They provide evidence that antibody maturation occurs within a dynamic multiway hub wherein transcription and mutation of different genes occur non-competitively.
dlvr.it
June 27, 2025 at 10:57 PM
Online Now: Regulation of somatic hypermutation by higher-order chromatin structure Online now:
Reposted
So cool, a novel archaeon with a super small genome and no metabolic genes!
"An unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses."
www.biorxiv.org/content/10.1...
"An unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses."
www.biorxiv.org/content/10.1...

A cellular entity retaining only its replicative core: Hidden archaeal lineage with an ultra-reduced genome
Defining the minimal genetic requirements for cellular life remains a fundamental question in biology. Genomic exploration continually reveals novel microbial lineages, often exhibiting extreme genome...
www.biorxiv.org
June 27, 2025 at 4:29 AM
So cool, a novel archaeon with a super small genome and no metabolic genes!
"An unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses."
www.biorxiv.org/content/10.1...
"An unprecedented level of metabolic dependence on a host, a condition that challenges the functional distinctions between minimal cellular life and viruses."
www.biorxiv.org/content/10.1...
Reposted
Many cells during development are exposed to caspase activation and yet survive. Why some die and not others ? We found out that the memory of previous caspase activation bias significantly later death comitment and bias death distribution and single cell decision
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
May 21, 2025 at 9:40 AM
Many cells during development are exposed to caspase activation and yet survive. Why some die and not others ? We found out that the memory of previous caspase activation bias significantly later death comitment and bias death distribution and single cell decision
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted
Out @nature.com: Clonal tracing with somatic epimutations
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
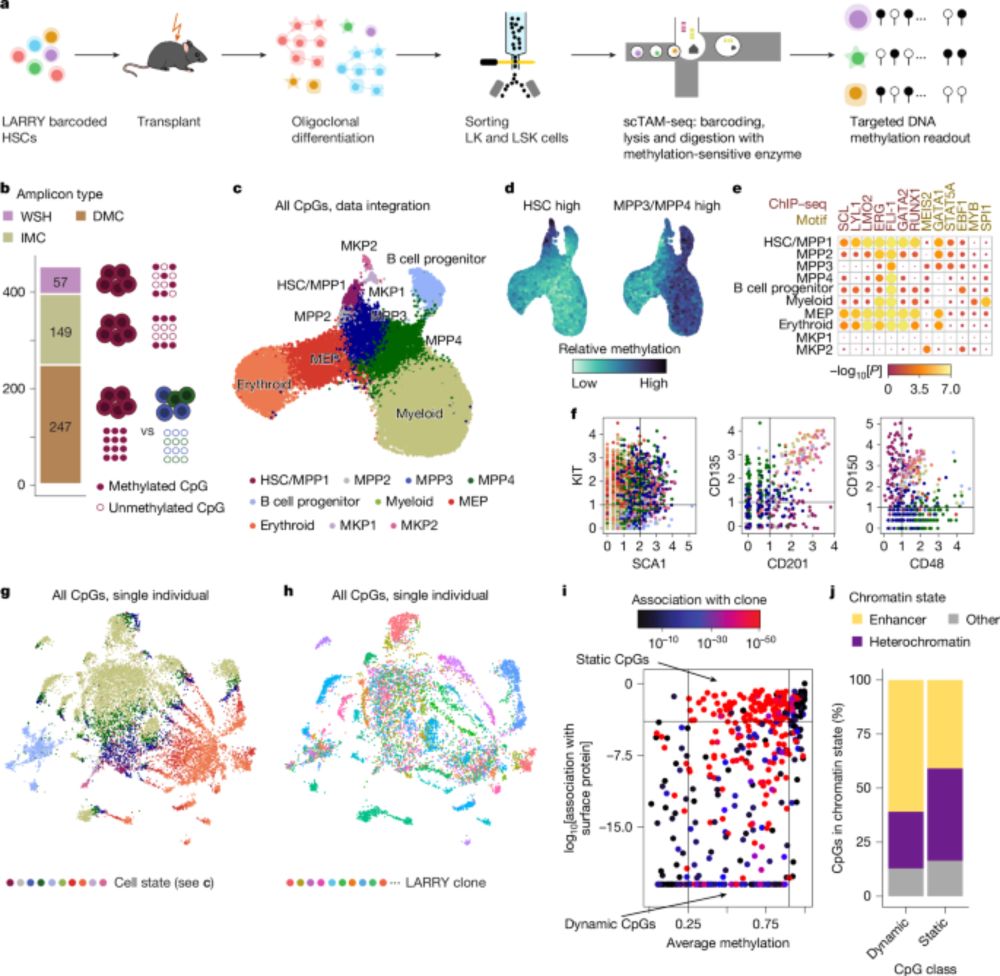
Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
May 21, 2025 at 3:44 PM
Out @nature.com: Clonal tracing with somatic epimutations
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
Reposted
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
www.cell.com
May 8, 2025 at 4:07 PM
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
Reposted
🐛🕷️🕸️ @science.org Hawaiian caterpillar patrols spiderwebs camouflaged in insect prey’s body parts | Science www.science.org/doi/10.1126/... #entomology
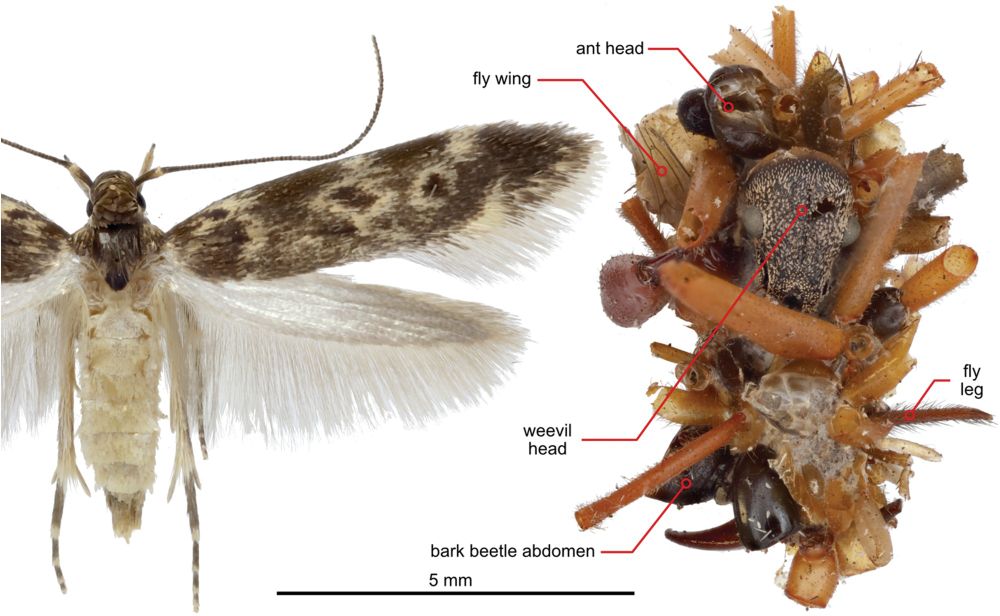

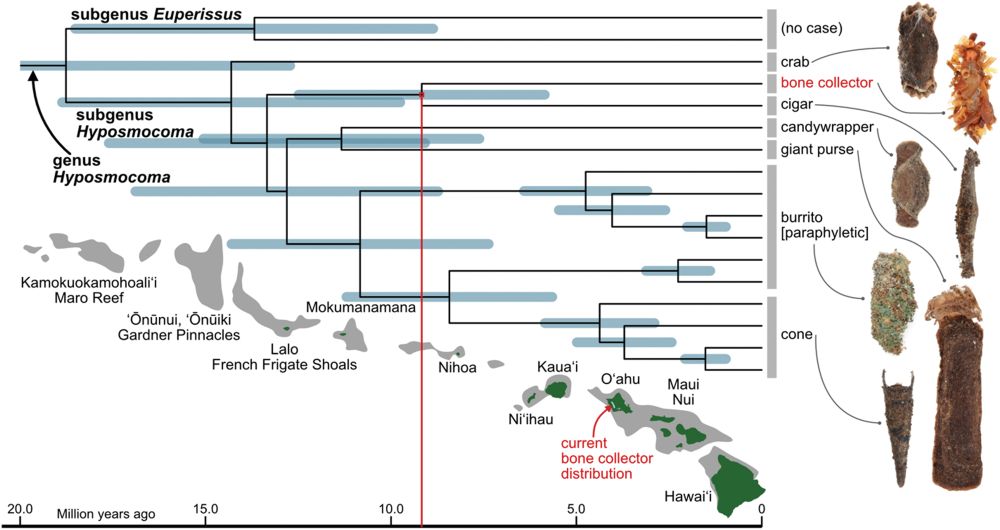
April 24, 2025 at 6:42 PM
🐛🕷️🕸️ @science.org Hawaiian caterpillar patrols spiderwebs camouflaged in insect prey’s body parts | Science www.science.org/doi/10.1126/... #entomology
Reposted
Nature research paper: DNA-guided transcription factor interactions extend human gene regulatory code
https://go.nature.com/4j13XRc
https://go.nature.com/4j13XRc
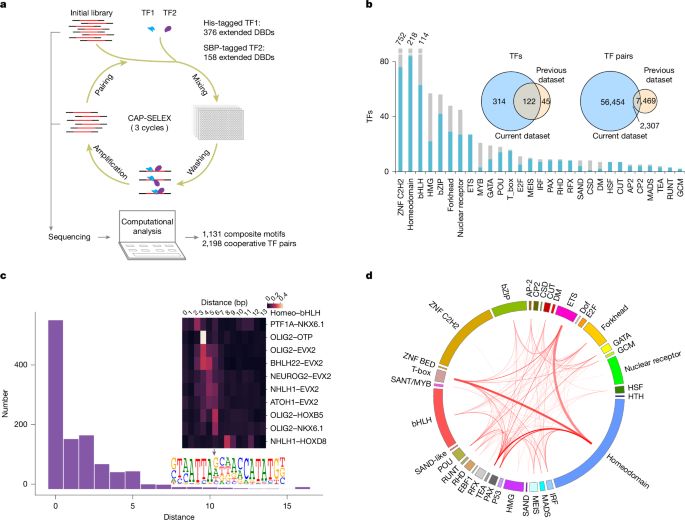
DNA-guided transcription factor interactions extend human gene regulatory code - Nature
A large-scale analysis of DNA-bound transcription factors (TFs) shows how the presence of DNA markedly affects the landscape of TF interactions, and identifies composite motifs that are recognized by complexes of TFs rather than by individual ones.
go.nature.com
April 9, 2025 at 5:24 PM
Nature research paper: DNA-guided transcription factor interactions extend human gene regulatory code
https://go.nature.com/4j13XRc
https://go.nature.com/4j13XRc
Reposted
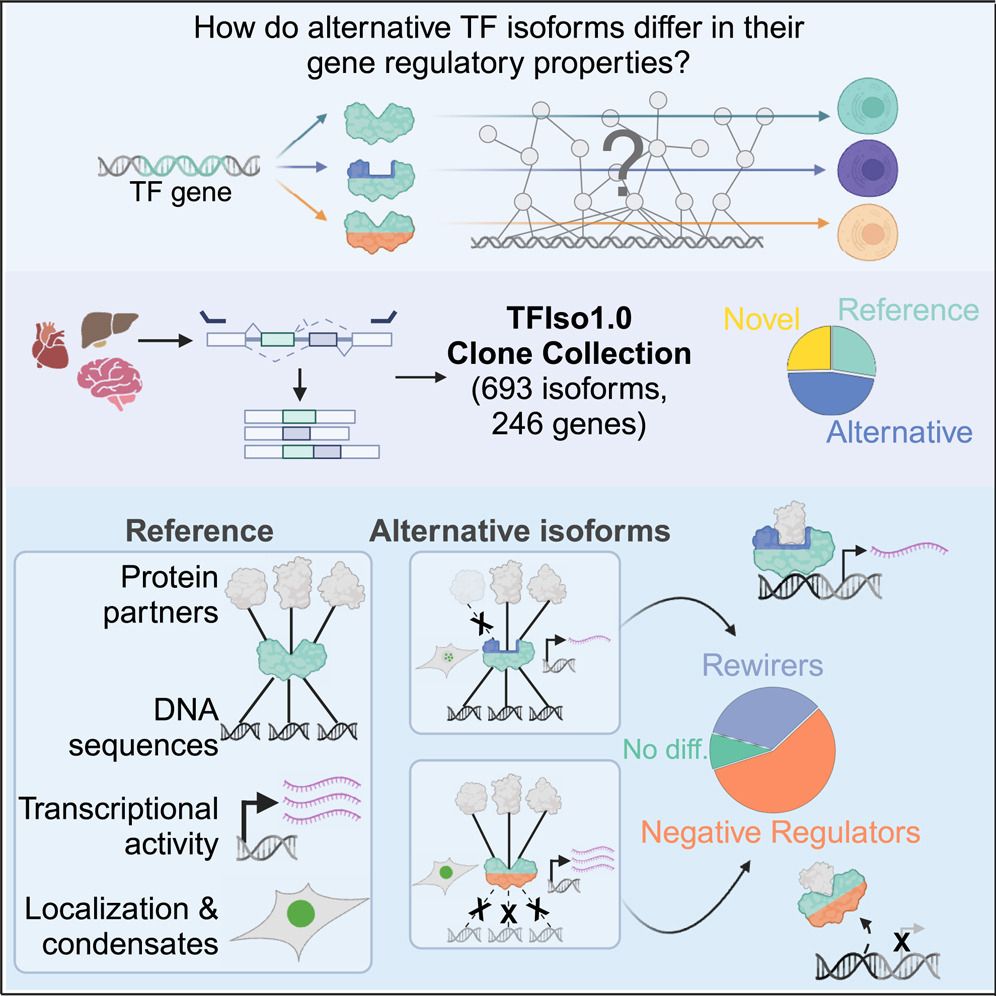
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
March 26, 2025 at 5:11 PM
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
Reposted
1/ H3K27me3 mimicry has repeatedly emerged through evolution, but what's the physiological relevance?
We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains.
🧵
www.biorxiv.org/content/10.1...
We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains.
🧵
www.biorxiv.org/content/10.1...
March 15, 2025 at 12:12 AM
1/ H3K27me3 mimicry has repeatedly emerged through evolution, but what's the physiological relevance?
We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains.
🧵
www.biorxiv.org/content/10.1...
We show that JARID2 and PALI1 mimic H3K27me3 to antagonise PRC2 in vivo and restrict the spread of Polycomb domains.
🧵
www.biorxiv.org/content/10.1...
Reposted
𝐇𝐨𝐰 𝐝𝐨 𝐜𝐞𝐥𝐥𝐬 𝐫𝐞𝐦𝐞𝐦𝐛𝐞𝐫 𝐰𝐡𝐨 𝐭𝐡𝐞𝐲 𝐚𝐫𝐞 𝐚𝐟𝐭𝐞𝐫 𝐃𝐍𝐀 𝐫𝐞𝐩𝐥𝐢𝐜𝐚𝐭𝐢𝐨𝐧? Our new study “Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability” is out in 𝘚𝘤𝘪𝘦𝘯𝘤𝘦 𝘈𝘥𝘷𝘢𝘯𝘤𝘦𝘴. Led by @lleonie.bsky.social @biranalva.bsky.social 🧵 More below👇
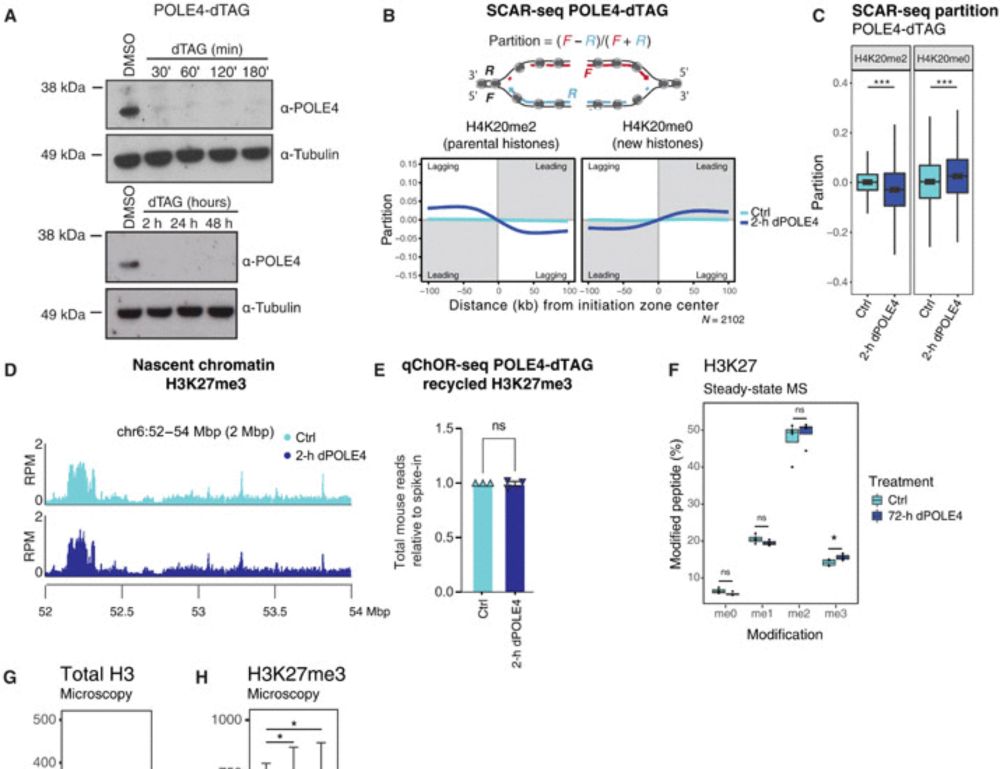
Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability
Losing parental histones during DNA replication fork passage challenges differentiation competence and cell viability.
tinyurl.com
February 19, 2025 at 7:35 PM
𝐇𝐨𝐰 𝐝𝐨 𝐜𝐞𝐥𝐥𝐬 𝐫𝐞𝐦𝐞𝐦𝐛𝐞𝐫 𝐰𝐡𝐨 𝐭𝐡𝐞𝐲 𝐚𝐫𝐞 𝐚𝐟𝐭𝐞𝐫 𝐃𝐍𝐀 𝐫𝐞𝐩𝐥𝐢𝐜𝐚𝐭𝐢𝐨𝐧? Our new study “Disabling leading and lagging strand histone transmission results in parental histones loss and reduced cell plasticity and viability” is out in 𝘚𝘤𝘪𝘦𝘯𝘤𝘦 𝘈𝘥𝘷𝘢𝘯𝘤𝘦𝘴. Led by @lleonie.bsky.social @biranalva.bsky.social 🧵 More below👇
Reposted
Yeah I don’t really care how they made the plasmids. Just give me the genbank file.
Can we make it a rule that when people publish papers involving generation of new plasmids, they put the plasmid sequence in the supplementary? Much more useful than "we digested plasmid A with enzyme B then ligated to C then digested with D then gel purified etc etc etc"
January 30, 2025 at 6:20 PM
Yeah I don’t really care how they made the plasmids. Just give me the genbank file.
Reposted
Not only does Bennu contain all 5 of the nucleobases that form DNA and RNA on Earth and 14 of the 20 amino acids found in known proteins, the asteroid’s amino acids hold a surprise
https://go.nature.com/4hDtGhL
https://go.nature.com/4hDtGhL

Asteroid fragments upend theory of how life on Earth bloomed
Samples from Bennu contain the chemical building blocks of life - but with a twist.
go.nature.com
January 29, 2025 at 4:38 PM
Not only does Bennu contain all 5 of the nucleobases that form DNA and RNA on Earth and 14 of the 20 amino acids found in known proteins, the asteroid’s amino acids hold a surprise
https://go.nature.com/4hDtGhL
https://go.nature.com/4hDtGhL
"Everything in epigenetics is circular". Here, we explore how memory that is stored locally on chromatin has a crucial role in stabilising cellular state, updating our view of the epigenetic landscape from one shaped by the genotype, to one moulded by experience
doi.org/10.1186/s130...
doi.org/10.1186/s130...

Chromatin-based memory as a self-stabilizing influence on cell identity - Genome Biology
Cell types are traditionally thought to be specified and stabilized by gene regulatory networks. Here, we explore how chromatin memory contributes to the specification and stabilization of cell states...
doi.org
December 30, 2024 at 10:58 PM
"Everything in epigenetics is circular". Here, we explore how memory that is stored locally on chromatin has a crucial role in stabilising cellular state, updating our view of the epigenetic landscape from one shaped by the genotype, to one moulded by experience
doi.org/10.1186/s130...
doi.org/10.1186/s130...
Reposted
📢Vote for the 2024 Development cover image of the year
@dev-journal.bsky.social featured 24 cover images in 2024. Now is the chance to pick your favourite!
Browse through the images and vote by 13 Jan:
https://thenode.biologists.com/vote-for-the-2024-development-cover-image-of-the-year/
#DevBio
@dev-journal.bsky.social featured 24 cover images in 2024. Now is the chance to pick your favourite!
Browse through the images and vote by 13 Jan:
https://thenode.biologists.com/vote-for-the-2024-development-cover-image-of-the-year/
#DevBio

December 27, 2024 at 1:56 PM
📢Vote for the 2024 Development cover image of the year
@dev-journal.bsky.social featured 24 cover images in 2024. Now is the chance to pick your favourite!
Browse through the images and vote by 13 Jan:
https://thenode.biologists.com/vote-for-the-2024-development-cover-image-of-the-year/
#DevBio
@dev-journal.bsky.social featured 24 cover images in 2024. Now is the chance to pick your favourite!
Browse through the images and vote by 13 Jan:
https://thenode.biologists.com/vote-for-the-2024-development-cover-image-of-the-year/
#DevBio
Reposted
Our latest: how to generate the notochord
"Timely TGFβ signalling inhibition induces notochord"
A thread:
www.nature.com/articles/s41...
"Timely TGFβ signalling inhibition induces notochord"
A thread:
www.nature.com/articles/s41...
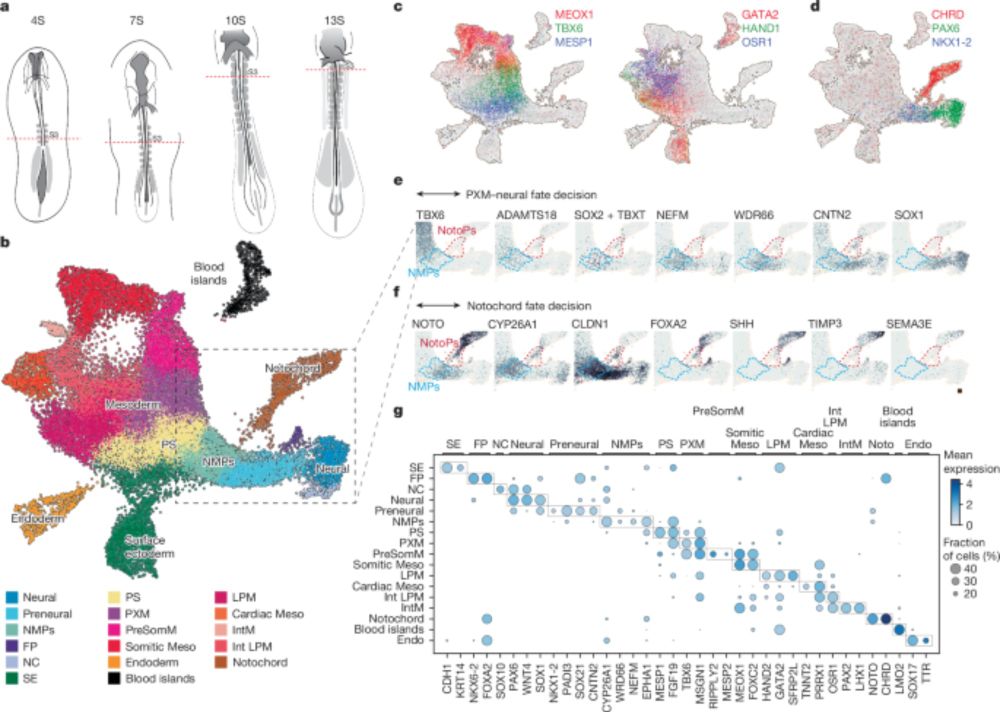
Timely TGFβ signalling inhibition induces notochord - Nature
Through analysis of developing chick embryos and in vitro differentiation of embryonic stem cells, a study develops a method to generate a model of the human trunk with a notochord.
www.nature.com
December 19, 2024 at 8:52 AM
Our latest: how to generate the notochord
"Timely TGFβ signalling inhibition induces notochord"
A thread:
www.nature.com/articles/s41...
"Timely TGFβ signalling inhibition induces notochord"
A thread:
www.nature.com/articles/s41...
Reposted
Crick researchers have recreated a tissue in the lab that acts like a developing embryo’s navigation system, directing cells where to build the spine and nervous system.
The findings were led by Tiago Rito with senior author @jamesbriscoe@bsky.social
www.crick.ac.uk/news/2024-12...
The findings were led by Tiago Rito with senior author @jamesbriscoe@bsky.social
www.crick.ac.uk/news/2024-12...

Building a backbone: scientists recreate the body’s ‘GPS system’ in the lab
Scientists at the Francis Crick Institute have generated human stem cell models1 which, for the first time, contain notochord – a tissue in the developing embryo that acts like a navigation system, di...
www.crick.ac.uk
December 18, 2024 at 5:24 PM
Crick researchers have recreated a tissue in the lab that acts like a developing embryo’s navigation system, directing cells where to build the spine and nervous system.
The findings were led by Tiago Rito with senior author @jamesbriscoe@bsky.social
www.crick.ac.uk/news/2024-12...
The findings were led by Tiago Rito with senior author @jamesbriscoe@bsky.social
www.crick.ac.uk/news/2024-12...
Reposted
New in J R Soc Interface: How temporal dynamics of morphogen signalling increases positional precision in vertebrate neural tube development
Combining Shh & BMP signals over time provides significantly more spatial information than static gradients alone
doi.org/10.1098/rsif... #DevBio
Combining Shh & BMP signals over time provides significantly more spatial information than static gradients alone
doi.org/10.1098/rsif... #DevBio

Dynamics of positional information in the vertebrate neural tube | Journal of The Royal Society Interface
In developing embryos, cells acquire distinct identities depending on their position
in a tissue. Secreted signalling molecules, known as morphogens, act as long-range
cues to provide the spatial info...
doi.org
December 13, 2024 at 10:56 AM
New in J R Soc Interface: How temporal dynamics of morphogen signalling increases positional precision in vertebrate neural tube development
Combining Shh & BMP signals over time provides significantly more spatial information than static gradients alone
doi.org/10.1098/rsif... #DevBio
Combining Shh & BMP signals over time provides significantly more spatial information than static gradients alone
doi.org/10.1098/rsif... #DevBio
Reposted
Excited to share that my main PhD project has been published! 🎉 We systematically designed and optimized reporters for 86 transcription factors in parallel. If you're interested in using these optimized reporters for your own research, don’t hesitate to reach out!

Optimized reporters for multiplexed detection of transcription factor activity
Direct measurements of transcription factor (TF) activity are crucial for understanding how TFs interpret signals and drive gene expression. TF reporter constructs have been widely used to detect activity in cell signaling, developmental biology, and disease models. However, many mammalian TFs lack reliable reporters. In this study, a library of reporters for 86 TFs was designed and evaluated to identify optimized “prime” TF reporters.
www.cell.com
December 13, 2024 at 8:49 AM
Excited to share that my main PhD project has been published! 🎉 We systematically designed and optimized reporters for 86 transcription factors in parallel. If you're interested in using these optimized reporters for your own research, don’t hesitate to reach out!



