🔗 doi.org/10.1093/molbev/msag011
#evobio #molbio #compbio

🔗 doi.org/10.1093/molbev/msag011
#evobio #molbio #compbio
link.springer.com/article/10.1...

link.springer.com/article/10.1...




doi.org/10.64898/202...
KIPEs3: Automatic annotation of biosynthesis pathways
doi.org/10.1101/2022...

doi.org/10.64898/202...
KIPEs3: Automatic annotation of biosynthesis pathways
doi.org/10.1101/2022...
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
Leverages deep learning to quickly and accurately assign KEGG Orthology, improving upon alignment-based methods for large datasets.




Leverages deep learning to quickly and accurately assign KEGG Orthology, improving upon alignment-based methods for large datasets.
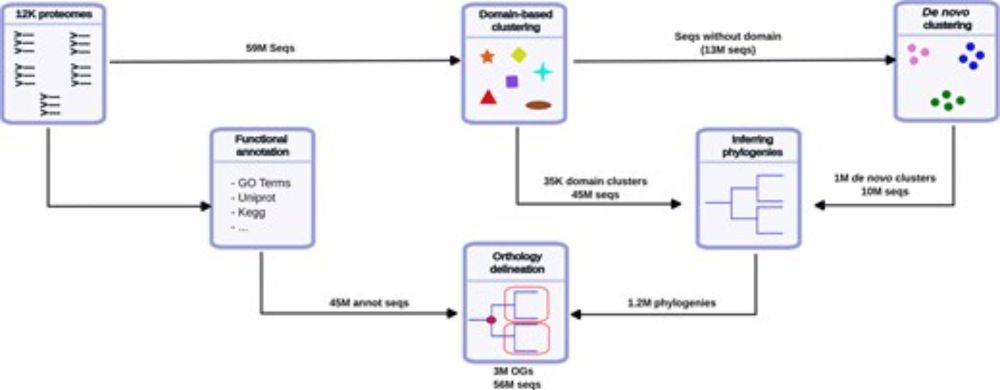



⬇️
academic.oup.com/nar/advance-...

⬇️
academic.oup.com/nar/advance-...





