@dnarnaresearch.bsky.social
When morphology stands still: constrained floral evolution in a mega diverse legume genus
doi.org/10.1101/2025...
Genetic factors explaining anthocyanin pigmentation differences
doi.org/10.1101/2023...
#Evolution #PlantSci
doi.org/10.1101/2025...
Genetic factors explaining anthocyanin pigmentation differences
doi.org/10.1101/2023...
#Evolution #PlantSci

When morphology stands still: constrained floral evolution in a mega diverse legume genus
· Changes in floral morphology are key for the evolutionary success of flowering plants. However, many species-rich and ecologically diverse groups show low variation in floral traits. Low morphologic...
doi.org
November 9, 2025 at 5:49 AM
When morphology stands still: constrained floral evolution in a mega diverse legume genus
doi.org/10.1101/2025...
Genetic factors explaining anthocyanin pigmentation differences
doi.org/10.1101/2023...
#Evolution #PlantSci
doi.org/10.1101/2025...
Genetic factors explaining anthocyanin pigmentation differences
doi.org/10.1101/2023...
#Evolution #PlantSci
Reposted
Friday Flower 009: Nigella arvensis ✨
Nigella’s flowers form a morphologically complex display with ornate bracts and blue sepals.
The true petals have diverse patterning with intricate, nectar-bearing structures that look alien.
Nigella’s flowers form a morphologically complex display with ornate bracts and blue sepals.
The true petals have diverse patterning with intricate, nectar-bearing structures that look alien.
November 7, 2025 at 9:17 AM
Friday Flower 009: Nigella arvensis ✨
Nigella’s flowers form a morphologically complex display with ornate bracts and blue sepals.
The true petals have diverse patterning with intricate, nectar-bearing structures that look alien.
Nigella’s flowers form a morphologically complex display with ornate bracts and blue sepals.
The true petals have diverse patterning with intricate, nectar-bearing structures that look alien.
Reposted
This is based on a talk I gave to the Crop Trust, before we conserve we need to understand the standing variation in the genebanks we have -- #ConservationGenomics kamounlab.medium.com/crispr-gene-...

November 7, 2025 at 4:57 PM
This is based on a talk I gave to the Crop Trust, before we conserve we need to understand the standing variation in the genebanks we have -- #ConservationGenomics kamounlab.medium.com/crispr-gene-...
Reposted
🌿🍇🍎 Espalier fruit trees are space-saving, beautiful, and productive! Train apples, pears, or figs flat against a wall or trellis for stunning architectural interest and easy harvesting. A perfect blend of art and horticulture! #Espalier #Gardening #FruitTrees

November 7, 2025 at 11:49 PM
🌿🍇🍎 Espalier fruit trees are space-saving, beautiful, and productive! Train apples, pears, or figs flat against a wall or trellis for stunning architectural interest and easy harvesting. A perfect blend of art and horticulture! #Espalier #Gardening #FruitTrees
Svirlpool: structural variant detection from long read sequencing by local assembly
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #Bioinformatics
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #Bioinformatics

Svirlpool: structural variant detection from long read sequencing by local assembly
Motivation: Long-Read Sequencing (LRS) promises great improvements in the detection of structural genome variants (SVs). However, existing methods are lacking in key areas such as the reliable detecti...
doi.org
November 8, 2025 at 6:42 AM
Svirlpool: structural variant detection from long read sequencing by local assembly
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #Bioinformatics
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #Bioinformatics
Nanoclustering of a plant transcription factor enables strong yet specific DNA binding
doi.org/10.1101/2025...
Automatic identification and annotation of MYB gene family members in plants
doi.org/10.1101/2021...
doi.org/10.1101/2025...
Automatic identification and annotation of MYB gene family members in plants
doi.org/10.1101/2021...

Nanoclustering of a plant transcription factor enables strong yet specific DNA binding
Transcription factors (TFs) are traditionally depicted as monomeric, or oligomeric, units that recognize and bind specific DNA sequences to regulate transcription. Emerging evidence suggests that many TFs can undergo liquid phase separation, resulting in condensates that bind DNA through a different mechanism. For the Auxin Response Factors (ARFs), canonical plant TFs, evidence for both scenarios exists. But which of these scenarios is operational in the plant nucleus is unclear. Here, we demonstrate using MpARF2 of Marchantia polymorpha that a third scenario is operational: MpARF2 forms nanoscopic clusters under physiological conditions. Nanoclusters combine DNA-binding features that cannot be accessed by the other two scenarios: high-affinity, switch-like, and sequence-specific. Our results suggest nanoclustering as a mechanism that equips TFs with the DNA-binding properties necessary for transcriptional regulation. ### Competing Interest Statement The authors have declared no competing interest. European Research Council, Catch 101000981 European Research Council, DIRNDL 833867 Human Frontiers Science Program, RGP0015/2022 Marie Skłodowska-Curie Individual Fellowship 2020, REOX 101026004 NWO, 184.036.012
doi.org
November 7, 2025 at 9:08 PM
Nanoclustering of a plant transcription factor enables strong yet specific DNA binding
doi.org/10.1101/2025...
Automatic identification and annotation of MYB gene family members in plants
doi.org/10.1101/2021...
doi.org/10.1101/2025...
Automatic identification and annotation of MYB gene family members in plants
doi.org/10.1101/2021...
Dynamic epigenetic and transcriptional regulatory network in pepper fruit development and ripening
doi.org/10.1101/2025...
Genome sequence and RNA-seq analysis reveal genetic basis of flower coloration in the giant water lily Victoria cruziana
doi.org/10.1101/2024...
doi.org/10.1101/2025...
Genome sequence and RNA-seq analysis reveal genetic basis of flower coloration in the giant water lily Victoria cruziana
doi.org/10.1101/2024...

Dynamic epigenetic and transcriptional regulatory network in pepper fruit development and ripening
Pepper (Capsicum) is among the most widely cultivated and consumed vegetable crops worldwide. Although extensive studies in model fruit crops such as tomato have provided insights into the genetic and...
doi.org
November 7, 2025 at 9:05 PM
Dynamic epigenetic and transcriptional regulatory network in pepper fruit development and ripening
doi.org/10.1101/2025...
Genome sequence and RNA-seq analysis reveal genetic basis of flower coloration in the giant water lily Victoria cruziana
doi.org/10.1101/2024...
doi.org/10.1101/2025...
Genome sequence and RNA-seq analysis reveal genetic basis of flower coloration in the giant water lily Victoria cruziana
doi.org/10.1101/2024...
Reposted
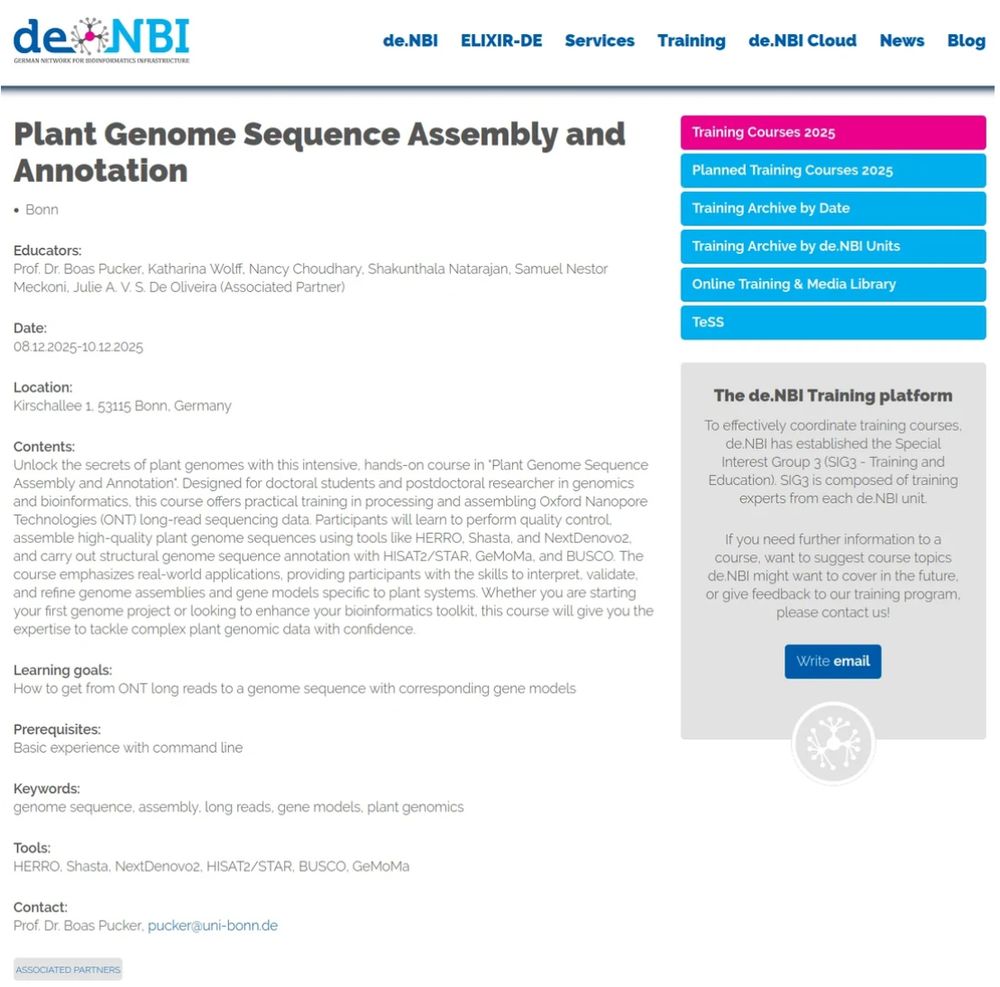
Curious about plant genomics? 🌿
Join our upcoming training courses to explore how plant genomes are assembled and annotated.
Details 👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #Bioinformatics #DeNBI #PlantScience
@denbi.bsky.social @puckerlab.bsky.social
Join our upcoming training courses to explore how plant genomes are assembled and annotated.
Details 👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #Bioinformatics #DeNBI #PlantScience
@denbi.bsky.social @puckerlab.bsky.social
November 6, 2025 at 5:25 PM
Curious about plant genomics? 🌿
Join our upcoming training courses to explore how plant genomes are assembled and annotated.
Details 👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #Bioinformatics #DeNBI #PlantScience
@denbi.bsky.social @puckerlab.bsky.social
Join our upcoming training courses to explore how plant genomes are assembled and annotated.
Details 👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #Bioinformatics #DeNBI #PlantScience
@denbi.bsky.social @puckerlab.bsky.social
Gene duplication dynamics and regulatory evolution shape the diversification of Asteraceae
doi.org/10.1101/2025...
DupyliCate - mining, classifying, and characterizing gene duplications
doi.org/10.1101/2025...
#Genomics #Bioinformatics #Evolution
doi.org/10.1101/2025...
DupyliCate - mining, classifying, and characterizing gene duplications
doi.org/10.1101/2025...
#Genomics #Bioinformatics #Evolution

Gene duplication dynamics and regulatory evolution shape the diversification of Asteraceae
The flowering plant order Asterales exhibits a striking disparity in species richness, with >30,000 species in Asteraceae compared to <50 in its sister family Calyceraceae. To investigate the genomic ...
doi.org
November 6, 2025 at 4:55 PM
Gene duplication dynamics and regulatory evolution shape the diversification of Asteraceae
doi.org/10.1101/2025...
DupyliCate - mining, classifying, and characterizing gene duplications
doi.org/10.1101/2025...
#Genomics #Bioinformatics #Evolution
doi.org/10.1101/2025...
DupyliCate - mining, classifying, and characterizing gene duplications
doi.org/10.1101/2025...
#Genomics #Bioinformatics #Evolution
Scaling up orphan crop research: A global genetic perspective of cowpea (Vigna unguiculata) diversity from 10,617 accessions
doi.org/10.1101/2025...
Reference-based QUantification Of gene Dispensability (QUOD)
doi.org/10.1101/2020...
#Genomics #Bioinformatics
doi.org/10.1101/2025...
Reference-based QUantification Of gene Dispensability (QUOD)
doi.org/10.1101/2020...
#Genomics #Bioinformatics

Scaling up orphan crop research: A global genetic perspective of cowpea (Vigna unguiculata) diversity from 10,617 accessions
Cowpea (Vigna unguiculata L. Walp) is a dryland legume crop, providing essential food and nutritional security for millions of people across the semi-arid tropics, in Africa, Asia, and Latin America. ...
doi.org
November 6, 2025 at 4:53 PM
Scaling up orphan crop research: A global genetic perspective of cowpea (Vigna unguiculata) diversity from 10,617 accessions
doi.org/10.1101/2025...
Reference-based QUantification Of gene Dispensability (QUOD)
doi.org/10.1101/2020...
#Genomics #Bioinformatics
doi.org/10.1101/2025...
Reference-based QUantification Of gene Dispensability (QUOD)
doi.org/10.1101/2020...
#Genomics #Bioinformatics
ProteoCast: a web server to predict, validate, and interpret missense variant effects
doi.org/10.1101/2025...
NAVIP: Unraveling the Influence of Neighboring Small Sequence Variants on Functional Impact Prediction
doi.org/10.1101/596718
doi.org/10.1101/2025...
NAVIP: Unraveling the Influence of Neighboring Small Sequence Variants on Functional Impact Prediction
doi.org/10.1101/596718

ProteoCast: a web server to predict, validate, and interpret missense variant effects
Understanding how mutations affect protein function remains critical yet challenging, particularly for variants in clinical databases lacking experimental characterisation and for intrinsically disord...
doi.org
November 5, 2025 at 5:40 AM
ProteoCast: a web server to predict, validate, and interpret missense variant effects
doi.org/10.1101/2025...
NAVIP: Unraveling the Influence of Neighboring Small Sequence Variants on Functional Impact Prediction
doi.org/10.1101/596718
doi.org/10.1101/2025...
NAVIP: Unraveling the Influence of Neighboring Small Sequence Variants on Functional Impact Prediction
doi.org/10.1101/596718
annoSnake: a Snakemake workflow for taxonomic and functional annotation of metagenomes and metagenome-assembled genomes (MAGs)
doi.org/10.1101/2025...
doi.org/10.1101/2025...

annoSnake: a Snakemake workflow for taxonomic and functional annotation of metagenomes and metagenome-assembled genomes (MAGs)
Omic technologies revolutionised research in microbiomics and helped decipher the complex community structure of gut microbiota, each comprising hundreds of species of bacteria, archaea, and protists. Even as sequencing costs decrease, understanding these communities remains a challenge. Here, we introduce annoSnake, a Snakemake workflow that facilitates taxonomic and functional annotation of metagenomes and metagenomic-assembled genomes (MAGs). This user-friendly and highly flexible workflow integrates state-of-the-art software packages, taking raw sequencing reads, assembling them into contigs, assigning taxonomies and annotating them using various popular protein databases. The workflow handles paired-end as well as interleaved read data, and provides publication-ready figures and tables for downstream analysis. annoSnake, as all Snakemake workflows, is highly scalable, reproducible, and portable, enabling execution across diverse high-performance computing environments with remarkable efficiency. ### Competing Interest Statement The authors have declared no competing interest.
doi.org
November 5, 2025 at 5:39 AM
annoSnake: a Snakemake workflow for taxonomic and functional annotation of metagenomes and metagenome-assembled genomes (MAGs)
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Reposted
Final step in the biosynthesis of iridoids elucidated
Iridoid-producing plants use a cyclase to synthesize the characteristic double-ring structure of this important class of defense compounds and medically relevant substances
buff.ly/kqtPHl2 via @maxplanck.de #PlantScience #Plants #Science
Iridoid-producing plants use a cyclase to synthesize the characteristic double-ring structure of this important class of defense compounds and medically relevant substances
buff.ly/kqtPHl2 via @maxplanck.de #PlantScience #Plants #Science

November 4, 2025 at 8:00 AM
Final step in the biosynthesis of iridoids elucidated
Iridoid-producing plants use a cyclase to synthesize the characteristic double-ring structure of this important class of defense compounds and medically relevant substances
buff.ly/kqtPHl2 via @maxplanck.de #PlantScience #Plants #Science
Iridoid-producing plants use a cyclase to synthesize the characteristic double-ring structure of this important class of defense compounds and medically relevant substances
buff.ly/kqtPHl2 via @maxplanck.de #PlantScience #Plants #Science
Reposted
🚀 de.NBI & ELIXIR-DE on tour at EOSC Symposium 2025 in Brussels!
Björn Grüning showcased the power of @galaxyproject.bsky.social Europe (usegalaxy.eu) and the de.NBI Cloud in the EOSC ecosystem.
#EOSC2025 #Bioinformatics #usegalaxy #ELIXIRDE #deNBI
Björn Grüning showcased the power of @galaxyproject.bsky.social Europe (usegalaxy.eu) and the de.NBI Cloud in the EOSC ecosystem.
#EOSC2025 #Bioinformatics #usegalaxy #ELIXIRDE #deNBI

November 4, 2025 at 8:55 AM
🚀 de.NBI & ELIXIR-DE on tour at EOSC Symposium 2025 in Brussels!
Björn Grüning showcased the power of @galaxyproject.bsky.social Europe (usegalaxy.eu) and the de.NBI Cloud in the EOSC ecosystem.
#EOSC2025 #Bioinformatics #usegalaxy #ELIXIRDE #deNBI
Björn Grüning showcased the power of @galaxyproject.bsky.social Europe (usegalaxy.eu) and the de.NBI Cloud in the EOSC ecosystem.
#EOSC2025 #Bioinformatics #usegalaxy #ELIXIRDE #deNBI
Reposted
Nature suggests you use their "Manuscript Adviser" bot to get advice before submitting
I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:
I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:

November 3, 2025 at 1:55 PM
Nature suggests you use their "Manuscript Adviser" bot to get advice before submitting
I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:
I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:
Reposted
🎉 We are excited to join this year's #Biohackathon Europe in Bad Saarow!
Looking forward to a week of fruitful projects, networking, & fun—thanks to the @elixir-europe.org Hub & everyone joining onsite or online!
🚀 @sneumann.bsky.social, Michael Crusoe, Nils Hoffmann & many more from across Europe.
Looking forward to a week of fruitful projects, networking, & fun—thanks to the @elixir-europe.org Hub & everyone joining onsite or online!
🚀 @sneumann.bsky.social, Michael Crusoe, Nils Hoffmann & many more from across Europe.

November 4, 2025 at 12:16 PM
🎉 We are excited to join this year's #Biohackathon Europe in Bad Saarow!
Looking forward to a week of fruitful projects, networking, & fun—thanks to the @elixir-europe.org Hub & everyone joining onsite or online!
🚀 @sneumann.bsky.social, Michael Crusoe, Nils Hoffmann & many more from across Europe.
Looking forward to a week of fruitful projects, networking, & fun—thanks to the @elixir-europe.org Hub & everyone joining onsite or online!
🚀 @sneumann.bsky.social, Michael Crusoe, Nils Hoffmann & many more from across Europe.
Reposted
🐢💙 Big win for sea turtle conservation!
Genome-informed conservation is helping keep these icons in our seas. 🌊
📄 Explore the 2025 publication: bit.ly/4ogC36B
🌊 Dive deeper into how these and other EBP-affiliated reference genomes are benefiting marine life in our latest newsletter: bit.ly/4qhJdZK
Genome-informed conservation is helping keep these icons in our seas. 🌊
📄 Explore the 2025 publication: bit.ly/4ogC36B
🌊 Dive deeper into how these and other EBP-affiliated reference genomes are benefiting marine life in our latest newsletter: bit.ly/4qhJdZK




November 4, 2025 at 5:16 PM
🐢💙 Big win for sea turtle conservation!
Genome-informed conservation is helping keep these icons in our seas. 🌊
📄 Explore the 2025 publication: bit.ly/4ogC36B
🌊 Dive deeper into how these and other EBP-affiliated reference genomes are benefiting marine life in our latest newsletter: bit.ly/4qhJdZK
Genome-informed conservation is helping keep these icons in our seas. 🌊
📄 Explore the 2025 publication: bit.ly/4ogC36B
🌊 Dive deeper into how these and other EBP-affiliated reference genomes are benefiting marine life in our latest newsletter: bit.ly/4qhJdZK
Symbiotic diversification relies on an ancestral gene network in plants
doi.org/10.1101/2025...
doi.org/10.1101/2025...

Symbiotic diversification relies on an ancestral gene network in plants
Symbioses have been fundamental to colonization of terrestrial ecosystems by plants and their evolution. Emergence of the ancient arbuscular mycorrhizal symbiosis was followed by the diversification o...
doi.org
November 4, 2025 at 5:46 PM
Symbiotic diversification relies on an ancestral gene network in plants
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Spatiotemporal regulation of arbuscular mycorrhizal symbiosis at cellular resolution
doi.org/10.1101/2025...
A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage
doi.org/10.1073/pnas...
doi.org/10.1101/2025...
A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage
doi.org/10.1073/pnas...

Spatiotemporal regulation of arbuscular mycorrhizal symbiosis at cellular resolution
Arbuscular mycorrhizal (AM) symbiosis develops through successive colonization of root epidermal and cortical cells, culminating in the formation of arbuscules, tree-like intracellular structures that...
doi.org
November 4, 2025 at 5:44 PM
Spatiotemporal regulation of arbuscular mycorrhizal symbiosis at cellular resolution
doi.org/10.1101/2025...
A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage
doi.org/10.1073/pnas...
doi.org/10.1101/2025...
A mycorrhiza-associated receptor-like kinase with an ancient origin in the green lineage
doi.org/10.1073/pnas...
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing
doi.org/10.1101/2025...
doi.org/10.1101/2025...

Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing
Nanopore direct RNA sequencing (DRS) coupled with Dorado modification-aware basecalling enables mapping of epitranscriptomic modifications including N6-methyladenosine (m6A) at the level of individual...
doi.org
November 3, 2025 at 5:49 AM
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Compound-specific DNA adduct profiling with nanopore sequencing and IonStats
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #LongReads
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #LongReads

Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
Background Experimental proof of gene function assignments in plants is based on mutant analyses. T-DNA insertion lines provided an invaluable resource of mutants and enabled systematic reverse geneti...
doi.org
November 2, 2025 at 7:02 AM
Compound-specific DNA adduct profiling with nanopore sequencing and IonStats
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #LongReads
doi.org/10.1101/2025...
Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis
doi.org/10.1101/2021...
#Genomics #LongReads
Effects of CO2 enrichment and environmental factors on photosynthesis, growth and yield and their interaction in cucumber
doi.org/10.1101/2025...
Large-scale Phylogenomics Reveals Systematic Loss of Anthocyanin Biosynthesis Genes at the Family Level in Cucurbitaceae
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Large-scale Phylogenomics Reveals Systematic Loss of Anthocyanin Biosynthesis Genes at the Family Level in Cucurbitaceae
doi.org/10.1101/2025...

Effects of carbon dioxide enrichment and environmental factors on photosynthesis, growth and yield and their interaction in cucumber: a meta-analysis
Despite the ongoing increase in atmospheric carbon dioxide concentrations, these levels remain well below the optimal threshold for cucumber growth and development. In addressing the challenges posed ...
doi.org
November 2, 2025 at 6:54 AM
Effects of CO2 enrichment and environmental factors on photosynthesis, growth and yield and their interaction in cucumber
doi.org/10.1101/2025...
Large-scale Phylogenomics Reveals Systematic Loss of Anthocyanin Biosynthesis Genes at the Family Level in Cucurbitaceae
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Large-scale Phylogenomics Reveals Systematic Loss of Anthocyanin Biosynthesis Genes at the Family Level in Cucurbitaceae
doi.org/10.1101/2025...
Diverse haplotypes at a complex Solanum americanum locus confer resistance to Phytophthora infestans and P. capsici
doi.org/10.1101/2025...
Phylogenomics and metabolic engineering reveal a conserved gene cluster in Solanaceae plants for withanolide biosynthesis
doi.org/10.1101/2024...
doi.org/10.1101/2025...
Phylogenomics and metabolic engineering reveal a conserved gene cluster in Solanaceae plants for withanolide biosynthesis
doi.org/10.1101/2024...

Diverse haplotypes at a complex Solanum americanum locus confer resistance to Phytophthora infestans and P. capsici
Plants encounter diverse pathogens and have evolved a two-layered innate immune system to detect pathogen molecules and activate defense mechanisms that restrict infection. Most cloned plant Resistanc...
doi.org
November 2, 2025 at 6:53 AM
Diverse haplotypes at a complex Solanum americanum locus confer resistance to Phytophthora infestans and P. capsici
doi.org/10.1101/2025...
Phylogenomics and metabolic engineering reveal a conserved gene cluster in Solanaceae plants for withanolide biosynthesis
doi.org/10.1101/2024...
doi.org/10.1101/2025...
Phylogenomics and metabolic engineering reveal a conserved gene cluster in Solanaceae plants for withanolide biosynthesis
doi.org/10.1101/2024...
Reposted
@igemhq.bsky.social and SB8 was unforgettable.
Would love to supervise a team on flower design for the new art/design village 🎨🌹🧬
Would love to supervise a team on flower design for the new art/design village 🎨🌹🧬
October 31, 2025 at 5:16 PM
@igemhq.bsky.social and SB8 was unforgettable.
Would love to supervise a team on flower design for the new art/design village 🎨🌹🧬
Would love to supervise a team on flower design for the new art/design village 🎨🌹🧬
Reposted
Interested in long-read plant genomics? 🌱
We are sharing our ONT P2 Solo sequencing outputs per flowcell — explore our real data and experience here:
👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #LongReads #PlantSci @puckerlab.bsky.social
We are sharing our ONT P2 Solo sequencing outputs per flowcell — explore our real data and experience here:
👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #LongReads #PlantSci @puckerlab.bsky.social

November 1, 2025 at 10:47 AM
Interested in long-read plant genomics? 🌱
We are sharing our ONT P2 Solo sequencing outputs per flowcell — explore our real data and experience here:
👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #LongReads #PlantSci @puckerlab.bsky.social
We are sharing our ONT P2 Solo sequencing outputs per flowcell — explore our real data and experience here:
👉 www.izmb.uni-bonn.de/en/pbb/news#...
#Genomics #LongReads #PlantSci @puckerlab.bsky.social

