Yasu Arimura
@yarimura.bsky.social
Elucidating the structural basis of biological events on chromosomes | chromatin, Xenopus egg extract, MagIC-cryo-EM | Assistant Professor at Fred Hutch https://research.fredhutch.org/arimura/en.html
@jhigginswriter.bsky.social wrote an article about my lab and our frog 🐸 at the Hutch! Thank you!
Dr. @yarimura.bsky.social brings frogs back to Fred Hutch after a long absence to study the structure of DNA-linked molecular complexes that change during the cell cycle and malfunction in cancer and other diseases. bit.ly/3J8S5Ae

Frogs help Fred Hutch find the shape of small things
Frogs have made significant contributions to Fred Hutch science over the years as a model organism that shares much of our biology, but they've been absent from the menagerie for about two decades. Dr...
bit.ly
October 29, 2025 at 6:29 AM
@jhigginswriter.bsky.social wrote an article about my lab and our frog 🐸 at the Hutch! Thank you!
Reposted by Yasu Arimura
Dr. @yarimura.bsky.social brings frogs back to Fred Hutch after a long absence to study the structure of DNA-linked molecular complexes that change during the cell cycle and malfunction in cancer and other diseases. bit.ly/3J8S5Ae

Frogs help Fred Hutch find the shape of small things
Frogs have made significant contributions to Fred Hutch science over the years as a model organism that shares much of our biology, but they've been absent from the menagerie for about two decades. Dr...
bit.ly
October 27, 2025 at 9:33 PM
Dr. @yarimura.bsky.social brings frogs back to Fred Hutch after a long absence to study the structure of DNA-linked molecular complexes that change during the cell cycle and malfunction in cancer and other diseases. bit.ly/3J8S5Ae
Reposted by Yasu Arimura
Looking for your next step in fly genetics? The Rajan Lab at Fred Hutch is hiring a staff scientist + postdoc for projects on fat–brain signals, immunity, and brain aging. Stable, long-term role in a collaborative lab. Apply: careers-fhcrc.icims.com/jobs/30062/j...

Fred Hutchinson Cancer Center
Fred Hutch is dedicated to the elimination of cancer and related diseases as causes of human suffering and death.
careers-fhcrc.icims.com
September 16, 2025 at 11:32 PM
Looking for your next step in fly genetics? The Rajan Lab at Fred Hutch is hiring a staff scientist + postdoc for projects on fat–brain signals, immunity, and brain aging. Stable, long-term role in a collaborative lab. Apply: careers-fhcrc.icims.com/jobs/30062/j...
Reposted by Yasu Arimura
My awesome Drosophila colleague Akhila Rajan at Fred Hutch (Seattle) is recruiting both a staff scientist and a postdoc to study fat–brain communication, innate immunity, mitochondrial signaling, and brain senescence. Great team, great environment. Apply here: careers-fhcrc.icims.com/jobs/30062/j...

Fred Hutchinson Cancer Center
Fred Hutch is dedicated to the elimination of cancer and related diseases as causes of human suffering and death.
careers-fhcrc.icims.com
September 17, 2025 at 1:05 AM
My awesome Drosophila colleague Akhila Rajan at Fred Hutch (Seattle) is recruiting both a staff scientist and a postdoc to study fat–brain communication, innate immunity, mitochondrial signaling, and brain senescence. Great team, great environment. Apply here: careers-fhcrc.icims.com/jobs/30062/j...
Reposted by Yasu Arimura
New online: Computational design of sequence-specific DNA-binding proteins

Computational design of sequence-specific DNA-binding proteins
Nature Structural & Molecular Biology, Published online: 12 September 2025; doi:10.1038/s41594-025-01669-4The authors develop a computational method to design small DNA-binding proteins (DBPs) that target specific sequences. Designed DBPs show structural accuracy and function in both bacterial and mammalian cells for transcriptional regulation.
go.nature.com
September 12, 2025 at 10:29 AM
New online: Computational design of sequence-specific DNA-binding proteins
Reposted by Yasu Arimura
One more movie!
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
August 29, 2025 at 12:36 AM
One more movie!
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
Reposted by Yasu Arimura
In new study led by @bdadonaite.bsky.social, we measure how spike mutations affect function & antigenicity of spike of KP.3.1.1 strain of SARS-CoV-2.
Sheds light on how key neutralizing epitopes are changing & importance of RBD up/down motion.
www.biorxiv.org/content/10.1...
Sheds light on how key neutralizing epitopes are changing & importance of RBD up/down motion.
www.biorxiv.org/content/10.1...

Spike mutations that affect the function and antigenicity of recent KP.3.1.1-like SARS-CoV-2 variants
SARS-CoV-2 is under strong evolutionary selection to acquire mutations in its spike protein that reduce neutralization by human polyclonal antibodies. Here we use pseudovirus-based deep mutational sca...
www.biorxiv.org
August 20, 2025 at 5:22 AM
In new study led by @bdadonaite.bsky.social, we measure how spike mutations affect function & antigenicity of spike of KP.3.1.1 strain of SARS-CoV-2.
Sheds light on how key neutralizing epitopes are changing & importance of RBD up/down motion.
www.biorxiv.org/content/10.1...
Sheds light on how key neutralizing epitopes are changing & importance of RBD up/down motion.
www.biorxiv.org/content/10.1...
Reposted by Yasu Arimura
WORK!
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍

August 13, 2025 at 1:30 PM
WORK!
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍
Reposted by Yasu Arimura
The peer-reviewed VOR for the MagIC-cryo-EM paper by @yarimura.bsky.social and @1001hak.bsky.social is finally published!
Congrats Yasu!
Congrats Yasu!
Researchers in the #FunabikiLab have devised a way to visualize molecules that are very rare, very small, or hard to produce naturally—including some viruses.
@elife.bsky.social #RockefellerScience
@elife.bsky.social #RockefellerScience

New method dramatically improves cryo-EM’s imaging capabilities - News
Researchers have devised a way to visualize molecules that are very rare, very small, or hard to produce naturally, including viruses.
www.rockefeller.edu
May 20, 2025 at 5:55 PM
The peer-reviewed VOR for the MagIC-cryo-EM paper by @yarimura.bsky.social and @1001hak.bsky.social is finally published!
Congrats Yasu!
Congrats Yasu!
Reposted by Yasu Arimura

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
May 6, 2025 at 3:08 PM

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
Reposted by Yasu Arimura
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
April 11, 2025 at 8:35 AM
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
Reposted by Yasu Arimura
Ever wondered what the lonely histone H3C110 does? We found that it’s lost in many fungi. Adding it back boosts histone H3 copper reductase activity, rescues iron defects and modulates lifespan, suggesting an evolutionary tradeoff between these effects.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
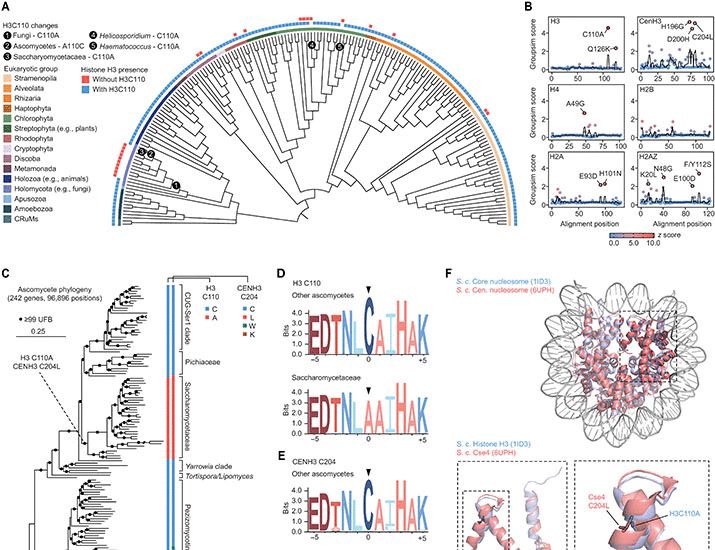
Histone H3 cysteine 110 enhances iron metabolism and modulates replicative life span in Saccharomyces cerevisiae
Histone H3 cysteine 110 enhances iron metabolism and modulates replicative life span in yeast.
www.science.org
April 11, 2025 at 9:43 PM
Ever wondered what the lonely histone H3C110 does? We found that it’s lost in many fungi. Adding it back boosts histone H3 copper reductase activity, rescues iron defects and modulates lifespan, suggesting an evolutionary tradeoff between these effects.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Reposted by Yasu Arimura
New preprint on 3D heterochromatin architecture in human cells! Great collab with @sergiocruzleon.bsky.social & @johannesbetz.bsky.social from @hummerlab.bsky.social, @marinalusic.bsky.social & the Turoňová lab. Many thanks to my supervisor @becklab.bsky.social. bioRxiv: tinyurl.com/3a74uanv 🧵👇
April 11, 2025 at 9:04 AM
New preprint on 3D heterochromatin architecture in human cells! Great collab with @sergiocruzleon.bsky.social & @johannesbetz.bsky.social from @hummerlab.bsky.social, @marinalusic.bsky.social & the Turoňová lab. Many thanks to my supervisor @becklab.bsky.social. bioRxiv: tinyurl.com/3a74uanv 🧵👇
Reposted by Yasu Arimura
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
April 6, 2025 at 9:45 AM
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
Reposted by Yasu Arimura
Our new paper is out@ScienceAdvances👇
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
March 29, 2025 at 1:06 AM
Our new paper is out@ScienceAdvances👇
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
Reposted by Yasu Arimura
We quantitatively measured nucleosome motion across the 4 chromatin classes, from euchromatin to heterochromatin.
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
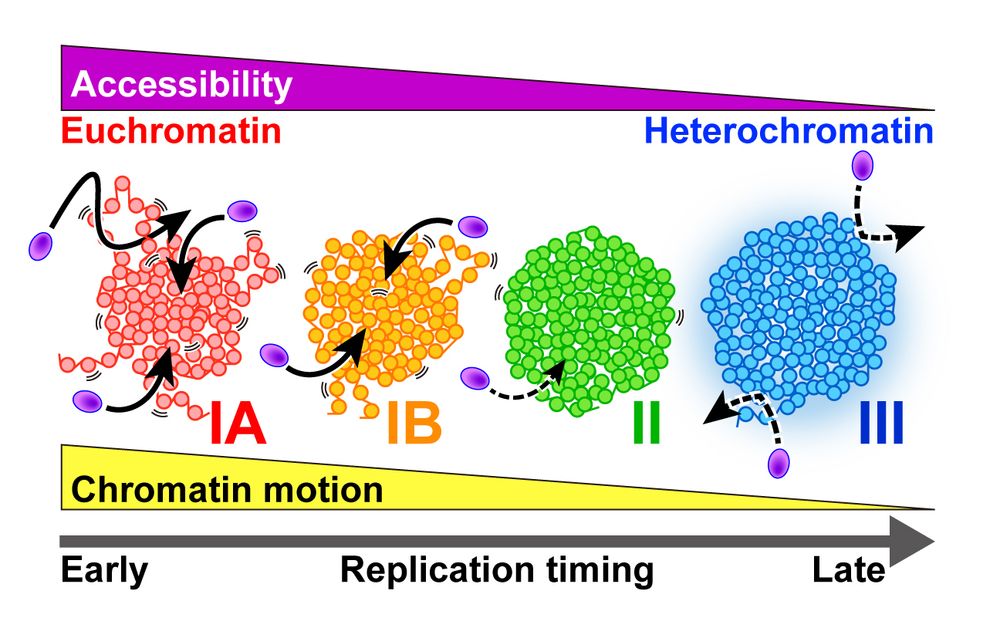
March 29, 2025 at 1:06 AM
We quantitatively measured nucleosome motion across the 4 chromatin classes, from euchromatin to heterochromatin.
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
Reposted by Yasu Arimura
Proud of this work, led by the amazing Jeannette Tenthorey (now Assistant Professor at UCSF), demonstrating the adaptive power of single indel mutations in host-virus arms races, but a little sad that this is the coda to her amazing postdoc in the Malik & Emerman labs.
www.cell.com/cell-genomic...
www.cell.com/cell-genomic...
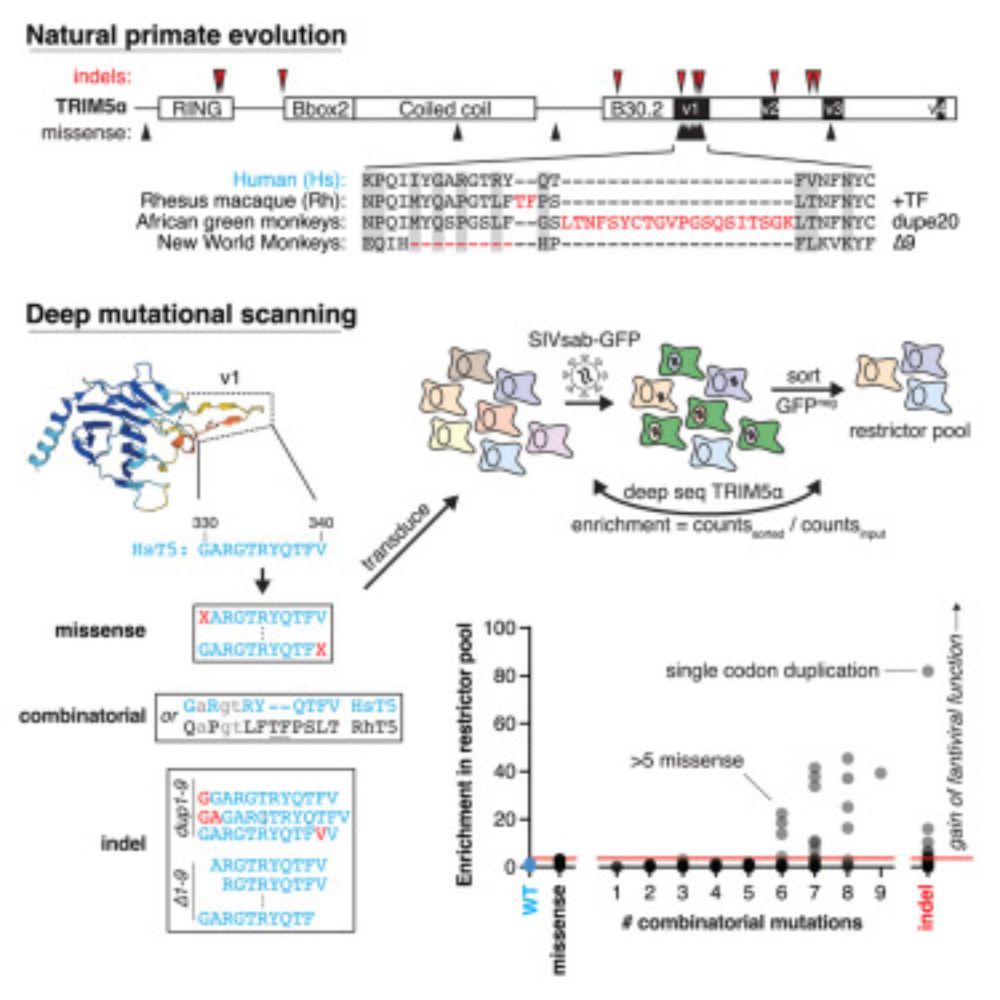
Indels allow antiviral proteins to evolve functional novelty inaccessible by missense mutations
Tenthorey et al. compare the effects of missense and indel mutations on the acquisition
of functional novelty by the rapidly evolving antiviral protein TRIM5α. They find
that single indel mutations al...
www.cell.com
March 27, 2025 at 9:36 PM
Proud of this work, led by the amazing Jeannette Tenthorey (now Assistant Professor at UCSF), demonstrating the adaptive power of single indel mutations in host-virus arms races, but a little sad that this is the coda to her amazing postdoc in the Malik & Emerman labs.
www.cell.com/cell-genomic...
www.cell.com/cell-genomic...
Reposted by Yasu Arimura
First post on bsky. We've updated our preprint about the change in the histone H3 variant on chromatin during Zygotic Genome Activation. Thanks to new experiments by Anusha, it seems like our initial, simplistic idea that the embryo "ran out" of H3 was not right.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Local nuclear to cytoplasmic ratio regulates H3.3 incorporation via cell cycle state during zygotic genome activation
Early embryos often have unique chromatin states prior to zygotic genome activation (ZGA). In Drosophila, ZGA occurs after 13 reductive nuclear divisions during which the nuclear to cytoplasmic (N/C) ...
www.biorxiv.org
December 13, 2024 at 5:21 PM
First post on bsky. We've updated our preprint about the change in the histone H3 variant on chromatin during Zygotic Genome Activation. Thanks to new experiments by Anusha, it seems like our initial, simplistic idea that the embryo "ran out" of H3 was not right.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Yasu Arimura
In a new Science study, cryo–electron tomography captures the in-cell architecture of the mitochondrial respiratory chain, illuminating how the coordinated action of molecular machines drives life’s fundamental energy conversion.
Learn more in this week's issue: scim.ag/3FA3Ygq
Learn more in this week's issue: scim.ag/3FA3Ygq
March 20, 2025 at 6:05 PM
In a new Science study, cryo–electron tomography captures the in-cell architecture of the mitochondrial respiratory chain, illuminating how the coordinated action of molecular machines drives life’s fundamental energy conversion.
Learn more in this week's issue: scim.ag/3FA3Ygq
Learn more in this week's issue: scim.ag/3FA3Ygq
Reposted by Yasu Arimura
Happy that Jon Chen @jonchenjk.bsky.social's cryo-ET and confocal study of RPE-1 chromatin in situ is online www.embopress.org/doi/full/10.1038/s44318-025-00407-2
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
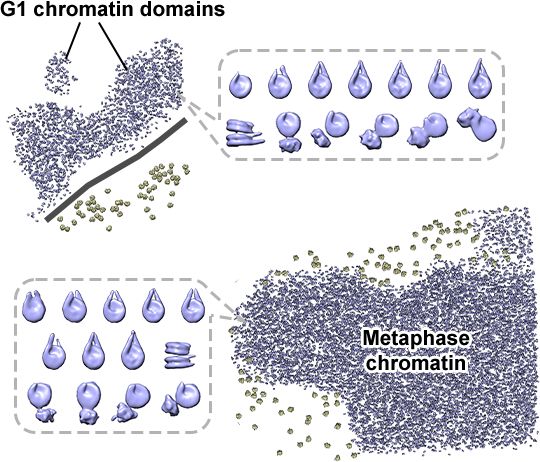
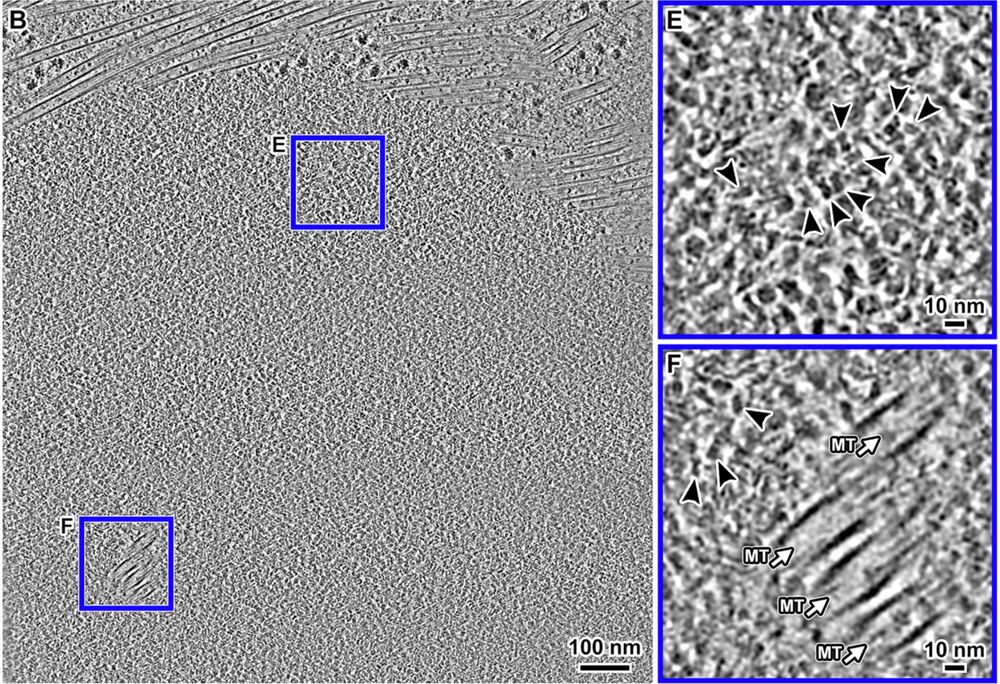
March 17, 2025 at 9:36 PM
Happy that Jon Chen @jonchenjk.bsky.social's cryo-ET and confocal study of RPE-1 chromatin in situ is online www.embopress.org/doi/full/10.1038/s44318-025-00407-2
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
Reposted by Yasu Arimura
Wonderful visit to @hutchbasicsci.bsky.social today and thanks to @yarimura.bsky.social for hosting and dinner with the always entertaining @suebiggins.bsky.social and @harmitmalik.bsky.social. A day discussing science with amazing people is unbeatable.

March 12, 2025 at 5:26 AM
Wonderful visit to @hutchbasicsci.bsky.social today and thanks to @yarimura.bsky.social for hosting and dinner with the always entertaining @suebiggins.bsky.social and @harmitmalik.bsky.social. A day discussing science with amazing people is unbeatable.
Reposted by Yasu Arimura
My first, first author paper is now on bioRxiv! 🤩
Linker histone H1 is a liquid-like "glue" condensing chromatin, which revises textbooks! 📖✨
biorxiv.org/content/10.1...
Huge thanks to my PI, @kazu-maeshima.bsky.social, for supervision.
Amazing collab with @rcollepardo.bsky.social’s group!" (1/n)
Linker histone H1 is a liquid-like "glue" condensing chromatin, which revises textbooks! 📖✨
biorxiv.org/content/10.1...
Huge thanks to my PI, @kazu-maeshima.bsky.social, for supervision.
Amazing collab with @rcollepardo.bsky.social’s group!" (1/n)
March 10, 2025 at 9:12 AM
My first, first author paper is now on bioRxiv! 🤩
Linker histone H1 is a liquid-like "glue" condensing chromatin, which revises textbooks! 📖✨
biorxiv.org/content/10.1...
Huge thanks to my PI, @kazu-maeshima.bsky.social, for supervision.
Amazing collab with @rcollepardo.bsky.social’s group!" (1/n)
Linker histone H1 is a liquid-like "glue" condensing chromatin, which revises textbooks! 📖✨
biorxiv.org/content/10.1...
Huge thanks to my PI, @kazu-maeshima.bsky.social, for supervision.
Amazing collab with @rcollepardo.bsky.social’s group!" (1/n)
Reposted by Yasu Arimura
I am super excited to share that my work at @yifan_ucsf has just been published in Nature🎉 From developing pYC KI system, it was a long journey to understand endogenous protein dynamics. I appreciate the support from FASN team! 🕺Let's dive into the endogenous & dynamic protein world🏊🏄🚣
March 2, 2025 at 10:25 AM
I am super excited to share that my work at @yifan_ucsf has just been published in Nature🎉 From developing pYC KI system, it was a long journey to understand endogenous protein dynamics. I appreciate the support from FASN team! 🕺Let's dive into the endogenous & dynamic protein world🏊🏄🚣
Reposted by Yasu Arimura
Researchers at Fred Hutch have discovered an overlooked mechanism driving aggressive breast and brain tumors involving genes so ancient — more than 2 billion years old — that they fly under the radar of standard genetic sequencing methods. 1/9 www.science.org/doi/10.1126/...

February 13, 2025 at 7:28 PM
Researchers at Fred Hutch have discovered an overlooked mechanism driving aggressive breast and brain tumors involving genes so ancient — more than 2 billion years old — that they fly under the radar of standard genetic sequencing methods. 1/9 www.science.org/doi/10.1126/...
Reposted by Yasu Arimura
Huge congratulations @jbloomlab.bsky.social!
Congrats to Fred Hutch evolutionary biologist
@jbloomlab.bsky.social on being named a fellow of the American Academy of Microbiology, an honorific leadership group within the American Society for Microbiology! Read more: bit.ly/3EFmnb1 @hutchbasicsci.bsky.social
@jbloomlab.bsky.social on being named a fellow of the American Academy of Microbiology, an honorific leadership group within the American Society for Microbiology! Read more: bit.ly/3EFmnb1 @hutchbasicsci.bsky.social

February 15, 2025 at 4:56 PM
Huge congratulations @jbloomlab.bsky.social!

