Tetsuya Yamada
@tyamadat.bsky.social
PhD student @UniHeidelberg | Kaessmann lab | UTokyo alumn
Pinned
Did transposable elements shape brain evolution — and if so, which ones, and in which cell states and lineages? Led by @tyamadat.bsky.social, we explored this question in cerebellum development using sequence-based deep learning models!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

So excited to see this out on bioRxiv! Huge thanks to @kaessmannlab.bsky.social and the amazing cerebellum team (@marisepp.bsky.social and @ioansarr.bsky.social) for all the support and insightful discussions that made this work possible!!
Reposted by Tetsuya Yamada
Excited to share Nona: a unifying multimodal masking framework for functional genomics.
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

November 10, 2025 at 9:01 PM
Excited to share Nona: a unifying multimodal masking framework for functional genomics.
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
Reposted by Tetsuya Yamada
Are you using any of our factor models, such as MOFA? 🛵
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

November 7, 2025 at 10:29 AM
Are you using any of our factor models, such as MOFA? 🛵
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n
Reposted by Tetsuya Yamada
Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

November 5, 2025 at 5:20 PM
Our latest paper has just been published in Cell!
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
Reposted by Tetsuya Yamada
🚀Excited to share our new preprint: “Programming human cell type-specific gene expression via an atlas of AI-designed enhancers”
We built the largest repository of AI-designed enhancers, validated across 10 human cell lines + mouse retina.
👉 www.biorxiv.org/content/10.1...
#AI #Genomics #SynBio
We built the largest repository of AI-designed enhancers, validated across 10 human cell lines + mouse retina.
👉 www.biorxiv.org/content/10.1...
#AI #Genomics #SynBio

Programming human cell type-specific gene expression via an atlas of AI-designed enhancers
Differentially active enhancers are key drivers of cell type specific gene expression. Active enhancers are found in open chromatin, which can be mapped at genome scale across tissue and cell types. T...
www.biorxiv.org
October 6, 2025 at 4:57 PM
🚀Excited to share our new preprint: “Programming human cell type-specific gene expression via an atlas of AI-designed enhancers”
We built the largest repository of AI-designed enhancers, validated across 10 human cell lines + mouse retina.
👉 www.biorxiv.org/content/10.1...
#AI #Genomics #SynBio
We built the largest repository of AI-designed enhancers, validated across 10 human cell lines + mouse retina.
👉 www.biorxiv.org/content/10.1...
#AI #Genomics #SynBio
Reposted by Tetsuya Yamada
New preprint led by the brilliant @aleksandra-marconi.bsky.social on cichlid brain diversification, fgf8a signalling and regulatory divergence with TEs on the mix! All part of a wonderful collaboration with @ebablab.bsky.social
www.biorxiv.org/content/10.1...
@camzoology.bsky.social
www.biorxiv.org/content/10.1...
@camzoology.bsky.social

October 24, 2025 at 8:45 PM
New preprint led by the brilliant @aleksandra-marconi.bsky.social on cichlid brain diversification, fgf8a signalling and regulatory divergence with TEs on the mix! All part of a wonderful collaboration with @ebablab.bsky.social
www.biorxiv.org/content/10.1...
@camzoology.bsky.social
www.biorxiv.org/content/10.1...
@camzoology.bsky.social
Reposted by Tetsuya Yamada
🧠🌟🐭 Excited to share some of my postdoc work on the evolution of dexterity!
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
October 22, 2025 at 8:41 PM
🧠🌟🐭 Excited to share some of my postdoc work on the evolution of dexterity!
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
So excited to see this out on bioRxiv! Huge thanks to @kaessmannlab.bsky.social and the amazing cerebellum team (@marisepp.bsky.social and @ioansarr.bsky.social) for all the support and insightful discussions that made this work possible!!
Did transposable elements shape brain evolution — and if so, which ones, and in which cell states and lineages? Led by @tyamadat.bsky.social, we explored this question in cerebellum development using sequence-based deep learning models!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

October 17, 2025 at 5:58 AM
So excited to see this out on bioRxiv! Huge thanks to @kaessmannlab.bsky.social and the amazing cerebellum team (@marisepp.bsky.social and @ioansarr.bsky.social) for all the support and insightful discussions that made this work possible!!
Reposted by Tetsuya Yamada
Excited to share the preprint from my main postdoc project! It’s been a long journey—huge thanks to everyone who made it possible, especially @leticiarm1618.bsky.social for being the best collaborator one could ask for, and the amazing @kaessmannlab.bsky.social lab for the invaluable support!
We are thrilled to share our new preprint entitled “The origin and molecular evolution of the mammalian liver cell architecture” www.biorxiv.org/content/10.1...
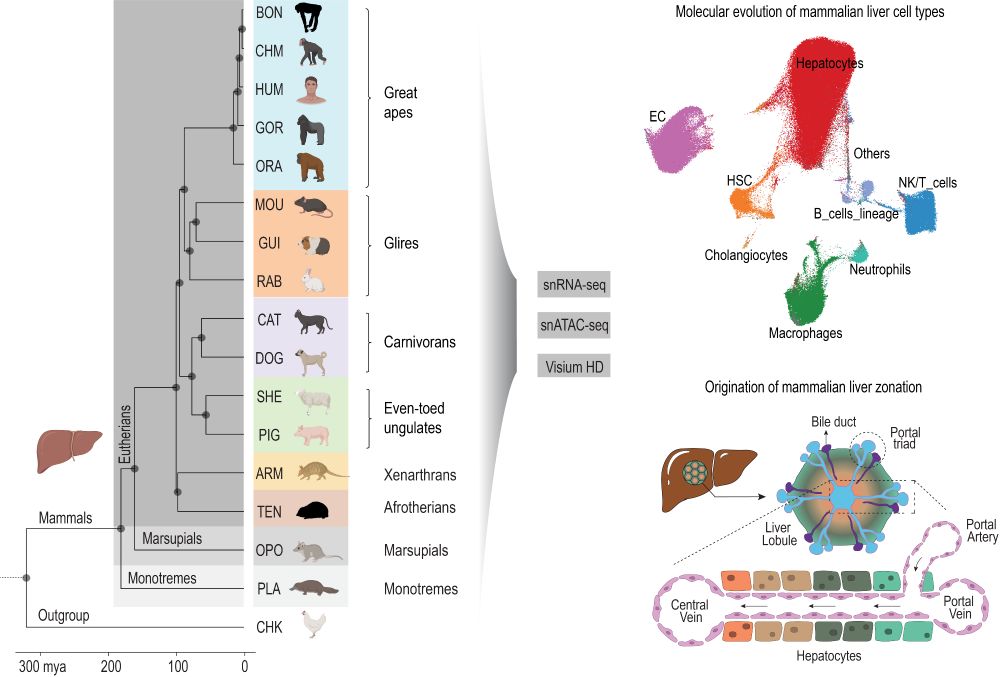
October 15, 2025 at 1:39 PM
Excited to share the preprint from my main postdoc project! It’s been a long journey—huge thanks to everyone who made it possible, especially @leticiarm1618.bsky.social for being the best collaborator one could ask for, and the amazing @kaessmannlab.bsky.social lab for the invaluable support!
Reposted by Tetsuya Yamada
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...

October 16, 2025 at 6:23 AM
🧠 The Lipid #Brain Atlas is out now! If you think #lipids are boring and membranes are all the same, prepare to be surprised. Led by @lucafusarbassini.bsky.social with Giovanni D'Angelo's lab, we mapped membrane lipids in the mouse brain at high resolution.
www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
Reposted by Tetsuya Yamada
We are thrilled to share our new preprint entitled “The origin and molecular evolution of the mammalian liver cell architecture” www.biorxiv.org/content/10.1...

October 15, 2025 at 12:51 PM
We are thrilled to share our new preprint entitled “The origin and molecular evolution of the mammalian liver cell architecture” www.biorxiv.org/content/10.1...
Reposted by Tetsuya Yamada
Which mutations rewire function of regulatory DNA?
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

October 9, 2025 at 12:03 PM
Which mutations rewire function of regulatory DNA?
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
Reposted by Tetsuya Yamada
Now published! Our paper on:
(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...

Sperm sequencing reveals extensive positive selection in the male germline - Nature
A combination of whole-genome NanoSeq with deep whole-exome and targeted NanoSeq is used to accurately characterize mutation rates and genes under positive selection in sperm cells.
www.nature.com
October 8, 2025 at 3:51 PM
Now published! Our paper on:
(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
Reposted by Tetsuya Yamada
New preprint from the lab! We discovered a transient fluidization in the basal region of human forebrains by tracking microdroplets in cerebral organoids.This “basal fluidization”, absent in gorilla and mouse, may contribute to greater surface expansion in human forebrains
1/
doi.org/10.1101/2025...
1/
doi.org/10.1101/2025...
October 8, 2025 at 1:05 PM
New preprint from the lab! We discovered a transient fluidization in the basal region of human forebrains by tracking microdroplets in cerebral organoids.This “basal fluidization”, absent in gorilla and mouse, may contribute to greater surface expansion in human forebrains
1/
doi.org/10.1101/2025...
1/
doi.org/10.1101/2025...
Reposted by Tetsuya Yamada
Happy to share work spearheaded by former grad student Colin Shew testing shared duplicated cis regulatory elements (CREs) using an MPRA. While we find some high effect CREs, collectively paralog differences represent modest effects accounting for observed gene expression divergence.
Influence of cis-regulatory elements on regulatory divergence in human segmental duplications https://www.biorxiv.org/content/10.1101/2025.10.03.680410v1
October 7, 2025 at 2:31 AM
Happy to share work spearheaded by former grad student Colin Shew testing shared duplicated cis regulatory elements (CREs) using an MPRA. While we find some high effect CREs, collectively paralog differences represent modest effects accounting for observed gene expression divergence.
Reposted by Tetsuya Yamada
Today in @nature.com, we present our work leveraging functional genomics and human blastoids to uncover a human-specific mechanism in preimplantation development driven by the endogenous retrovirus HERVK.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD

A human-specific regulatory mechanism revealed in a pre-implantation model
Nature - Genetic manipulation of blastoids reveals the role of recently emerged transposable elements and genes in human development.
rdcu.be
October 1, 2025 at 6:08 PM
Today in @nature.com, we present our work leveraging functional genomics and human blastoids to uncover a human-specific mechanism in preimplantation development driven by the endogenous retrovirus HERVK.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
Reposted by Tetsuya Yamada
New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...

September 26, 2025 at 6:06 PM
New paper from my lab and @jshendure.bsky.social lab! Led by the brilliant @zukailiu.bsky.social and @cxqiu.bsky.social. We tackled how anterior and posterior progenitor cells cooperate to self-organize into an embryonic structure (termed AP-gastruloid). (1/n) www.biorxiv.org/content/10.1...
Reposted by Tetsuya Yamada
We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
www.biorxiv.org/content/10.1...
(1/n)

September 22, 2025 at 5:29 AM
We are excited to share GPN-Star, a cost-effective, biologically grounded genomic language modeling framework that achieves state-of-the-art performance across a wide range of variant effect prediction tasks relevant to human genetics.
www.biorxiv.org/content/10.1...
(1/n)
www.biorxiv.org/content/10.1...
(1/n)
Reposted by Tetsuya Yamada
New preprint from our lab!
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/

September 19, 2025 at 3:03 AM
New preprint from our lab!
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/
What can we learn about the properties of gene regulatory elements by CRISPR’ing a random set of accessible sites in human cells?
Find out here: www.biorxiv.org/content/10.1...
👇
1/
Reposted by Tetsuya Yamada
Ever wondered how robust cellular identity is to external perturbations? Here we disrupt cellular environmnent in vivo and in vitro, and find cell population specific sensitivities. Environment sculpts development yes, but not all cells are made of the same wood. www.science.org/doi/10.1126/...

Cell-extrinsic controls over neocortical neuron fate and diversity
Cell-extrinsic cues are key for neocortical cell identity and diversity.
www.science.org
September 19, 2025 at 6:04 AM
Ever wondered how robust cellular identity is to external perturbations? Here we disrupt cellular environmnent in vivo and in vitro, and find cell population specific sensitivities. Environment sculpts development yes, but not all cells are made of the same wood. www.science.org/doi/10.1126/...
Reposted by Tetsuya Yamada
New from the lab: Fatty Acid Metabolism Sets the Species-specific Tempo of Cortical Development through Protein Acetylation Dynamics. Led by amazing trio: @ryo2iwata.bsky.social , Isabel Gallego, @emirerkol.bsky.social, together with @steinaerts.bsky.social , @fendtlab.bsky.social

Species-specific Rates of Fatty Acid Metabolism Set the Scale of Temporal Patterning of Corticogenesis through Protein Acetylation Dynamics
Developmental processes display temporal differences across species, leading to divergence in organ size and composition. In the cerebral cortex, neurons of diverse identities are generated sequential...
www.biorxiv.org
September 3, 2025 at 12:19 PM
New from the lab: Fatty Acid Metabolism Sets the Species-specific Tempo of Cortical Development through Protein Acetylation Dynamics. Led by amazing trio: @ryo2iwata.bsky.social , Isabel Gallego, @emirerkol.bsky.social, together with @steinaerts.bsky.social , @fendtlab.bsky.social
Reposted by Tetsuya Yamada
🧠🦈Excited to present our latest work🧠🦈Interested in brain evolution? And shark embryos? Then read on… Our work sheds light on the deep origins of our brain’s most complex regions.
September 2, 2025 at 5:51 PM
🧠🦈Excited to present our latest work🧠🦈Interested in brain evolution? And shark embryos? Then read on… Our work sheds light on the deep origins of our brain’s most complex regions.
Reposted by Tetsuya Yamada
Now published @natecoevo.nature.com with @annika-nichols.bsky.social, our latest on the evolution of 𝘴𝘭𝘦𝘦𝘱 across 𝟲𝟬 𝘀𝗽𝗲𝗰𝗶𝗲𝘀 of cichlid fishes! doi.org/10.1038/s415...
with members of the @schierlab.bsky.social and Salzburger labs, as well as the burgeoning Shafer lab @uoftcellsysbiol.bsky.social
with members of the @schierlab.bsky.social and Salzburger labs, as well as the burgeoning Shafer lab @uoftcellsysbiol.bsky.social
August 28, 2025 at 2:18 PM
Now published @natecoevo.nature.com with @annika-nichols.bsky.social, our latest on the evolution of 𝘴𝘭𝘦𝘦𝘱 across 𝟲𝟬 𝘀𝗽𝗲𝗰𝗶𝗲𝘀 of cichlid fishes! doi.org/10.1038/s415...
with members of the @schierlab.bsky.social and Salzburger labs, as well as the burgeoning Shafer lab @uoftcellsysbiol.bsky.social
with members of the @schierlab.bsky.social and Salzburger labs, as well as the burgeoning Shafer lab @uoftcellsysbiol.bsky.social
Reposted by Tetsuya Yamada
1) I am delighted to present this terrific tour de force research conducted by my post-doc Dr. Gayani Senevirathne @gayani.bsky.social and published today in Nature -
www.nature.com/articles/s41...
www.nature.com/articles/s41...

The evolution of hominin bipedalism in two steps - Nature
The human pelvis exhibits distinct spatiotemporal ossification patterns and an ilium cartilage growth plate that is shifted perpendicularly compared with those of other mammals and non-human primates—...
www.nature.com
August 27, 2025 at 3:13 PM
1) I am delighted to present this terrific tour de force research conducted by my post-doc Dr. Gayani Senevirathne @gayani.bsky.social and published today in Nature -
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Tetsuya Yamada
In the genomics community, we have focused pretty heavily on achieving state-of-the-art predictive performance.
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
August 27, 2025 at 4:20 PM
In the genomics community, we have focused pretty heavily on achieving state-of-the-art predictive performance.
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
While undoubtedly important, how we *use* these models after training is potentially even more important.
tangermeme v1.0.0 is out now. Hope you find it useful!
Reposted by Tetsuya Yamada
Excited to share our latest work on brain evolution, where we dive into the evolution of Cajal-Retzius cells! If you are interested in cell type evolution and cerebral cortex evo-devo, please read on… www.biorxiv.org/content/10.1... 🧵 1/7

Evolution of Cajal-Retzius Cells in Vertebrates from an Ancient Class of Tp73+ Neurons
In the developing cerebral cortex, Cajal Retzius (CR) cells are early-born neurons that orchestrate the development of mammalian-specific cortical features. However, this cell type has not been conclu...
www.biorxiv.org
August 25, 2025 at 7:43 PM
Excited to share our latest work on brain evolution, where we dive into the evolution of Cajal-Retzius cells! If you are interested in cell type evolution and cerebral cortex evo-devo, please read on… www.biorxiv.org/content/10.1... 🧵 1/7

