
Samuel Sledzieski
@samsl.io
Research Fellow @flatironinstitute.org @simonsfoundation.org
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
Pinned
Samuel Sledzieski
@samsl.io
· Jun 23
www.biorxiv.org
New preprint 🚨--protein language models + MD training ➡️ allosteric networks!
@sonyahanson.bsky.social and I are developing RocketSHP 🚀 for rapid genome-scale inference of local+correlated fluctuations + structure token distributions!
📄: www.biorxiv.org/content/10.1...
💻: github.com/flatironinst...
@sonyahanson.bsky.social and I are developing RocketSHP 🚀 for rapid genome-scale inference of local+correlated fluctuations + structure token distributions!
📄: www.biorxiv.org/content/10.1...
💻: github.com/flatironinst...
Reposted by Samuel Sledzieski
Announcing cryo-EM heterogeneity challenge #2, now dubbed the 2025 Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms (CAHRA)! Join us for a webinar next Friday (Nov 14th) to learn more. Datasets already posted here: heterogeneity.notion.site/challenge #cryoem

CAHRA 2025: Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms
A community-wide data processing challenge for cryo-electron microscopy.
heterogeneity.notion.site
November 7, 2025 at 10:45 PM
Announcing cryo-EM heterogeneity challenge #2, now dubbed the 2025 Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms (CAHRA)! Join us for a webinar next Friday (Nov 14th) to learn more. Datasets already posted here: heterogeneity.notion.site/challenge #cryoem
Reposted by Samuel Sledzieski
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
October 22, 2025 at 2:55 PM
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
D-SCRIPT + BMPI is out today in Bioinformatics!
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
October 12, 2025 at 5:20 PM
D-SCRIPT + BMPI is out today in Bioinformatics!
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
Reposted by Samuel Sledzieski
Integrative modelling of biomolecular dynamics
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...

October 2, 2025 at 7:54 AM
Integrative modelling of biomolecular dynamics
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...
Reposted by Samuel Sledzieski
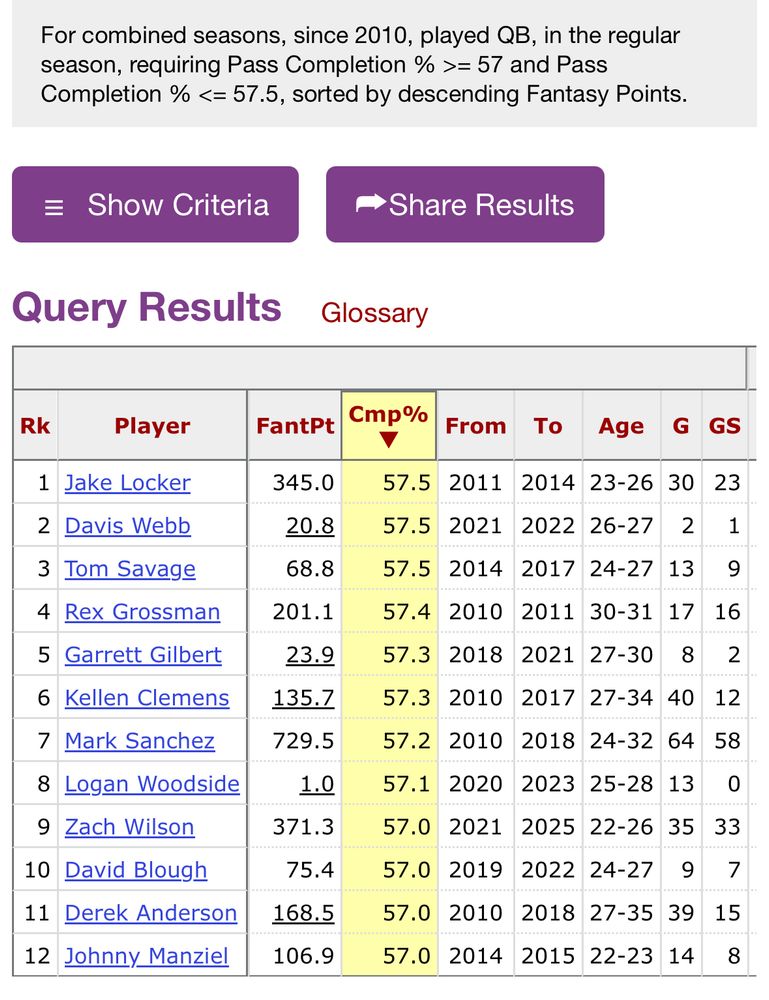
These are the quarterbacks since 2010 who complete passes at roughly the rate Microsoft Excel’s AI mode accurately edits spreadsheets
October 2, 2025 at 12:26 AM
These are the quarterbacks since 2010 who complete passes at roughly the rate Microsoft Excel’s AI mode accurately edits spreadsheets
Reposted by Samuel Sledzieski
*Job Alert*: postdoc positions available @flatironinstitute.org in Computational Mathematics: apply.interfolio.com/173401
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
apply.interfolio.com
September 19, 2025 at 10:40 AM
*Job Alert*: postdoc positions available @flatironinstitute.org in Computational Mathematics: apply.interfolio.com/173401
Reposted by Samuel Sledzieski
Reposted by Samuel Sledzieski
YouTube link for MLCB2025 is up! Starting in 30 min. www.youtube.com/live/19I7xTh...

Machine Learning in Computational Biology 2025
YouTube video by Machine Learning in Computational Biology
www.youtube.com
September 10, 2025 at 1:01 PM
YouTube link for MLCB2025 is up! Starting in 30 min. www.youtube.com/live/19I7xTh...
I’ll be at MLCB today and tomorrow, and talking about our latest work on modeling protein dynamics 🚀 tomorrow afternoon!
2025 Machine Learning in Computational Biology (#MLCB) meeting starts TODAY (9/10) at 9:30a (EST) at the NY Genome Center in NYC!
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf

MLCB - Schedule
The in-person component will be held at the New York Genome Center, 101 6th Ave, New York, NY 10013. All times below are Eastern Time.
mlcb.org
September 10, 2025 at 1:04 PM
I’ll be at MLCB today and tomorrow, and talking about our latest work on modeling protein dynamics 🚀 tomorrow afternoon!
Reposted by Samuel Sledzieski
2025 Machine Learning in Computational Biology (#MLCB) meeting starts TODAY (9/10) at 9:30a (EST) at the NY Genome Center in NYC!
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf

MLCB - Schedule
The in-person component will be held at the New York Genome Center, 101 6th Ave, New York, NY 10013. All times below are Eastern Time.
mlcb.org
September 10, 2025 at 11:42 AM
2025 Machine Learning in Computational Biology (#MLCB) meeting starts TODAY (9/10) at 9:30a (EST) at the NY Genome Center in NYC!
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf
We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf
Reposted by Samuel Sledzieski
Excited to share our recent work: Sparse autoencoders uncover biologically interpretable features in protein language model representations now in PNAS. Thread below 🧵

September 3, 2025 at 4:50 PM
Excited to share our recent work: Sparse autoencoders uncover biologically interpretable features in protein language model representations now in PNAS. Thread below 🧵
Reposted by Samuel Sledzieski
Announcing the Diffuse Project! We're unlocking protein dynamics through diffuse X-ray scattering - the overlooked signal that could revolutionize how we understand protein motion. seemay.substack.com/p/from-syste...
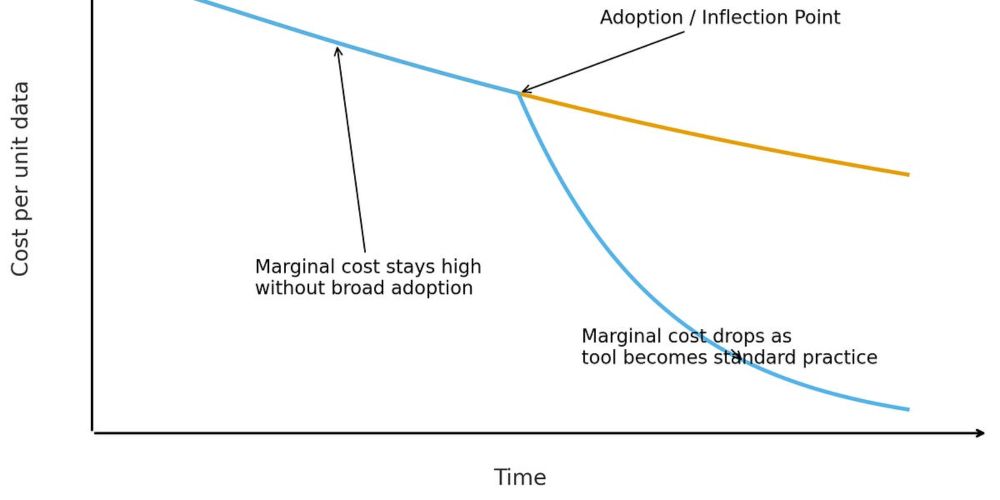
From systems operators to systems architects
Going up a level from data generation to think about the data systems we design and embed
seemay.substack.com
August 12, 2025 at 4:21 PM
Announcing the Diffuse Project! We're unlocking protein dynamics through diffuse X-ray scattering - the overlooked signal that could revolutionize how we understand protein motion. seemay.substack.com/p/from-syste...
Reposted by Samuel Sledzieski
The results of the ISCB Leadership vote are in!
Terms for the ISCB President-Elect and ISCB Vice President will start on January 21, 2026.
Terms for the ISCB President-Elect and ISCB Vice President will start on January 21, 2026.

August 13, 2025 at 1:08 PM
The results of the ISCB Leadership vote are in!
Terms for the ISCB President-Elect and ISCB Vice President will start on January 21, 2026.
Terms for the ISCB President-Elect and ISCB Vice President will start on January 21, 2026.
Reposted by Samuel Sledzieski
I just got the notice that all the FlyBase people at Harvard, including me, will be laid off on October 12. I'm devastated.
August 11, 2025 at 6:12 PM
I just got the notice that all the FlyBase people at Harvard, including me, will be laid off on October 12. I'm devastated.
Reposted by Samuel Sledzieski
We have a new preprint out on the Masala software suite, a free and open-source platform for easy biomolecular modelling methods development. www.biorxiv.org/content/10.1...

The open-source Masala software suite: Facilitating rapid methods development for synthetic heteropolymer design
Although canonical protein design has benefited from machine learning methods trained on databases of protein sequences and structures, synthetic heteropolymer design still relies heavily on physics-b...
www.biorxiv.org
July 14, 2025 at 1:07 PM
We have a new preprint out on the Masala software suite, a free and open-source platform for easy biomolecular modelling methods development. www.biorxiv.org/content/10.1...
Reposted by Samuel Sledzieski
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
August 5, 2025 at 6:31 AM
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
Reposted by Samuel Sledzieski
We are excited to share our preprint describing DirectContacts2! Here we develop a machine learning model to discriminate between direct and indirect protein interactions. We use our model to construct a highly accurate wiring diagram of the human cell.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

DirectContacts2: A network of direct physical protein interactions derived from high-throughput mass spectrometry experiments
Cellular function is driven by the activity proteins in stable complexes. Protein complex assembly depends on the direct physical association of component proteins. Advances in macromolecular structur...
www.biorxiv.org
July 29, 2025 at 8:58 PM
We are excited to share our preprint describing DirectContacts2! Here we develop a machine learning model to discriminate between direct and indirect protein interactions. We use our model to construct a highly accurate wiring diagram of the human cell.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Samuel Sledzieski
Stay tuned for details on the 6th edition of MLSB, officially happening this December in downtown San Diego, CA!
The MLSB workshop will be in San Diego, CA (co-located with NeurIPS) this year for its 6th edition in December 🧬🔬
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟

July 28, 2025 at 3:41 PM
Stay tuned for details on the 6th edition of MLSB, officially happening this December in downtown San Diego, CA!
Reposted by Samuel Sledzieski
That's a wrap! The results of the first #cryoEM heterogeneity challenge are up on biorxiv!
biorxiv.org/content/10.110
biorxiv.org/content/10.110
biorxiv.org
July 23, 2025 at 9:43 PM
That's a wrap! The results of the first #cryoEM heterogeneity challenge are up on biorxiv!
biorxiv.org/content/10.110
biorxiv.org/content/10.110
Reposted by Samuel Sledzieski
#FlatironCCB scientists Natalie Sauerwald, Olga Troyanskaya and colleagues leveraged @sparkforautism.bsky.social data to reveal four distinct groups that link autism-related traits with underlying genetics. Read more: www.simonsfoundation.org/2025/07/09/n... #science #biology #autismresearch

New Study Reveals Subclasses of Autism by Linking Traits to Genetics
New Study Reveals Subclasses of Autism by Linking Traits to Genetics on Simons Foundation
www.simonsfoundation.org
July 21, 2025 at 2:22 PM
#FlatironCCB scientists Natalie Sauerwald, Olga Troyanskaya and colleagues leveraged @sparkforautism.bsky.social data to reveal four distinct groups that link autism-related traits with underlying genetics. Read more: www.simonsfoundation.org/2025/07/09/n... #science #biology #autismresearch
Today we released a major new update to D-SCRIPT! 🚨
TL;DR, whole-proteome protein-protein interaction prediction is now significantly faster *and* less memory intensive 🚅 🧠
You can get started with the new version with `pip install dscript==0.3.0`
💻 github.com/samsledje/D-...
🧵⬇️
TL;DR, whole-proteome protein-protein interaction prediction is now significantly faster *and* less memory intensive 🚅 🧠
You can get started with the new version with `pip install dscript==0.3.0`
💻 github.com/samsledje/D-...
🧵⬇️
Release v0.3.0: 2025-07-22 -- Blocked, Multi-GPU Parallel Inference (BMPI) · samsledje/D-SCRIPT
What's Changed
Update evaluate.py and predict.py by @samsledje in #76
Modernization of D-SCRIPT in anticipation of v0.3.0 by @samsledje in #77
Update README.md by @samsledje in #78
Parallel, multi...
github.com
July 22, 2025 at 6:39 PM
Today we released a major new update to D-SCRIPT! 🚨
TL;DR, whole-proteome protein-protein interaction prediction is now significantly faster *and* less memory intensive 🚅 🧠
You can get started with the new version with `pip install dscript==0.3.0`
💻 github.com/samsledje/D-...
🧵⬇️
TL;DR, whole-proteome protein-protein interaction prediction is now significantly faster *and* less memory intensive 🚅 🧠
You can get started with the new version with `pip install dscript==0.3.0`
💻 github.com/samsledje/D-...
🧵⬇️
Reposted by Samuel Sledzieski
The short film "Molecular Duet" brings the dynamic world of microtubules to life. A collaboration between @flatironinstitute.org researcher Mahsa Mofidi and filmmaker Anne Sofie Nørskov. Watch in full: www.simonsfoundation.org/2025/07/16/w... #science #biology #film #FlatironCCB
July 17, 2025 at 8:13 PM
The short film "Molecular Duet" brings the dynamic world of microtubules to life. A collaboration between @flatironinstitute.org researcher Mahsa Mofidi and filmmaker Anne Sofie Nørskov. Watch in full: www.simonsfoundation.org/2025/07/16/w... #science #biology #film #FlatironCCB
Reposted by Samuel Sledzieski
From #FlatironCCB scientist Bargeen Turzo and filmmaker Grace Zhang, "How to Connect Two Bodies" is an experimental documentary exploring the invisible choreography in meaningful connections of objects across various scales. Watch in full: www.simonsfoundation.org/2025/07/09/w... #science #film
July 16, 2025 at 4:14 PM
From #FlatironCCB scientist Bargeen Turzo and filmmaker Grace Zhang, "How to Connect Two Bodies" is an experimental documentary exploring the invisible choreography in meaningful connections of objects across various scales. Watch in full: www.simonsfoundation.org/2025/07/09/w... #science #film
Reposted by Samuel Sledzieski
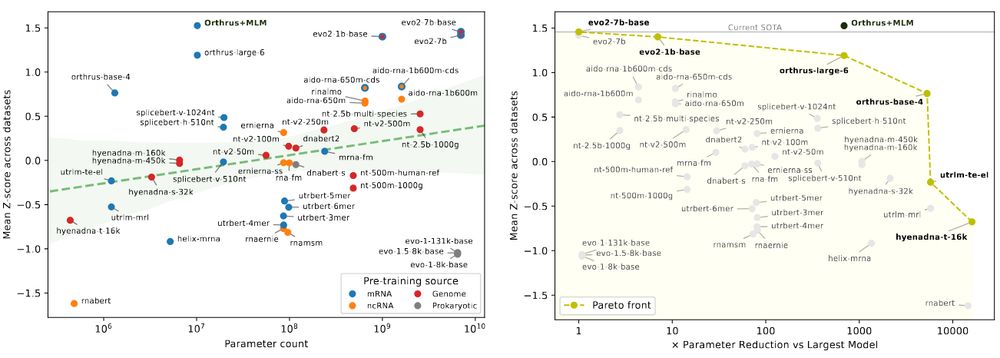
We're excited to release 𝐦𝐑𝐍𝐀𝐁𝐞𝐧𝐜𝐡, a new benchmark suite for mRNA biology containing 10 diverse datasets with 59 prediction tasks, evaluating 18 foundation model families.
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
July 15, 2025 at 6:41 PM
We're excited to release 𝐦𝐑𝐍𝐀𝐁𝐞𝐧𝐜𝐡, a new benchmark suite for mRNA biology containing 10 diverse datasets with 59 prediction tasks, evaluating 18 foundation model families.
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench


