
Samuel Sledzieski
@samsl.io
Research Fellow @flatironinstitute.org @simonsfoundation.org
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
We hope this update will significantly expand the types of researchers + compute systems that can use D-SCRIPT! Manuscript with benchmarks + implementation details coming hopefully soon, and thanks again so much to Daniel for his hard work pushing this out!
July 22, 2025 at 6:39 PM
We hope this update will significantly expand the types of researchers + compute systems that can use D-SCRIPT! Manuscript with benchmarks + implementation details coming hopefully soon, and thanks again so much to Daniel for his hard work pushing this out!
Along with a bunch of other quality-of-life updates (including a mode specifically designed for host x pathogen/symbiont interactions! 🦠), v0.3.0 will power the sorts of comparative genomics and network analyses that we imagined when we first designed D-SCRIPT.
July 22, 2025 at 6:39 PM
Along with a bunch of other quality-of-life updates (including a mode specifically designed for host x pathogen/symbiont interactions! 🦠), v0.3.0 will power the sorts of comparative genomics and network analyses that we imagined when we first designed D-SCRIPT.
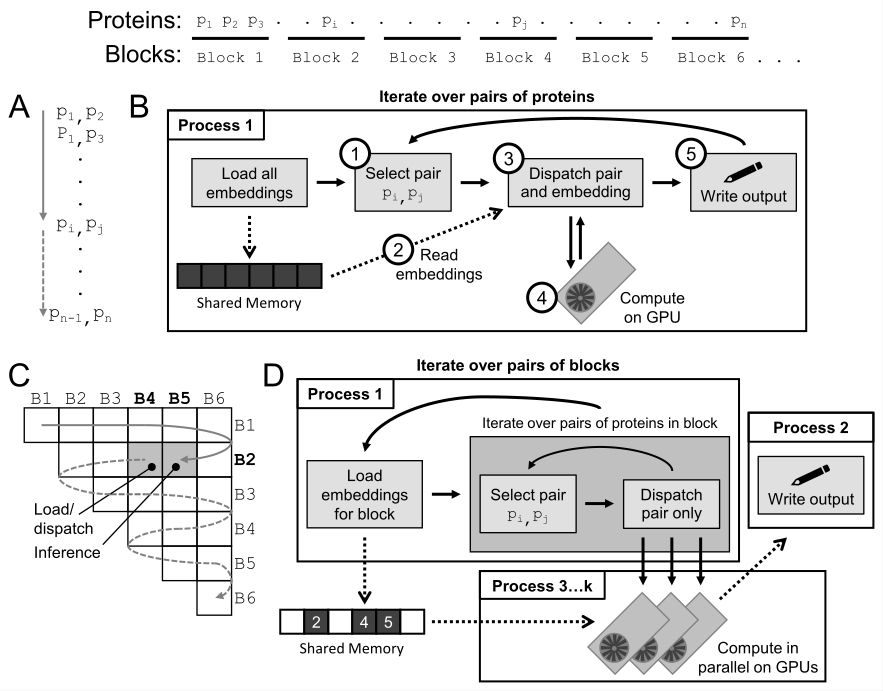
BMPI makes D-SCRIPT fully parallel in both embedding loading and inference, and can distribute inference to an arbitrary number of available GPUs. At the same time, the blocked inference procedure means that memory usage can be decreased arbitrarily low (with a small trade-off in speed).
July 22, 2025 at 6:39 PM
BMPI makes D-SCRIPT fully parallel in both embedding loading and inference, and can distribute inference to an arbitrary number of available GPUs. At the same time, the blocked inference procedure means that memory usage can be decreased arbitrarily low (with a small trade-off in speed).
In v0.3.0, we solve *both* of these issues, enabling efficient inference on both personal computers and multi-GPU HPC systems. The secret? Our new Blocked Multi-GPU Parallel Inference (BMPI) procedure, led by Daniel Schaffer (github.com/schafferde).
July 22, 2025 at 6:39 PM
In v0.3.0, we solve *both* of these issues, enabling efficient inference on both personal computers and multi-GPU HPC systems. The secret? Our new Blocked Multi-GPU Parallel Inference (BMPI) procedure, led by Daniel Schaffer (github.com/schafferde).

