
Samuel Sledzieski
@samsl.io
Research Fellow @flatironinstitute.org @simonsfoundation.org
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻
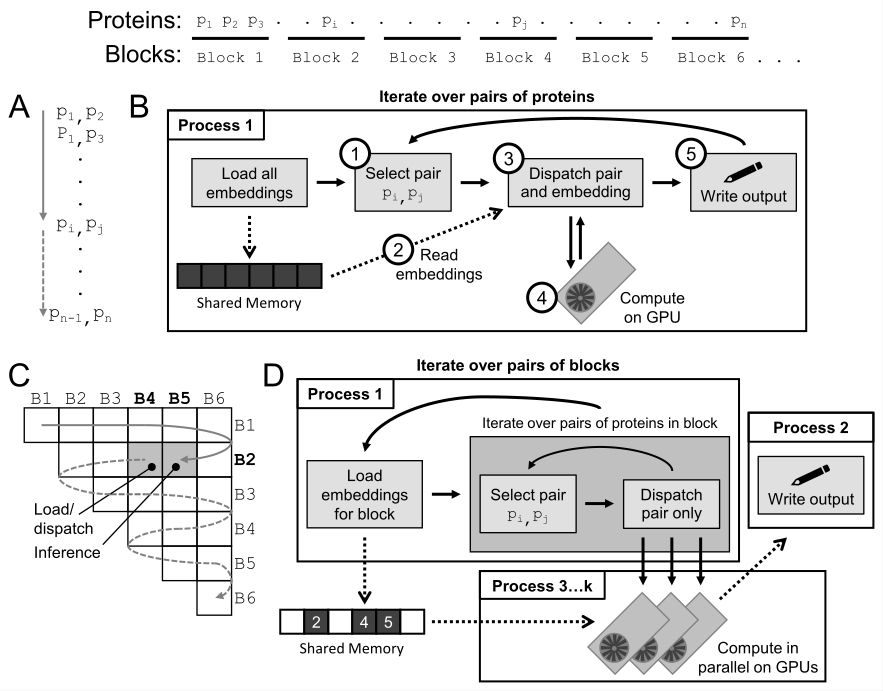
In v0.3.0, we solve *both* of these issues, enabling efficient inference on both personal computers and multi-GPU HPC systems. The secret? Our new Blocked Multi-GPU Parallel Inference (BMPI) procedure, led by Daniel Schaffer (github.com/schafferde).
July 22, 2025 at 6:39 PM
In v0.3.0, we solve *both* of these issues, enabling efficient inference on both personal computers and multi-GPU HPC systems. The secret? Our new Blocked Multi-GPU Parallel Inference (BMPI) procedure, led by Daniel Schaffer (github.com/schafferde).
Our key hypothesis is that these motions carry information about allosteric networks, despite not explicitly measuring larger conformational change. Inspired by terrific work from Federica Maschietto, we show that in silico predictions of these networks closely matches DMS results in KRAS.

June 23, 2025 at 8:42 PM
Our key hypothesis is that these motions carry information about allosteric networks, despite not explicitly measuring larger conformational change. Inspired by terrific work from Federica Maschietto, we show that in silico predictions of these networks closely matches DMS results in KRAS.
And we think we can! Thanks to excellent data curation efforts from @hkws.bsky.social, we show that RocketSHP-predicted fluctuations correlate well with experimental hetNOE measurements that capture fast + local movement.
![(a) Scatter plot showing RocketSHP predicted RMSF and hetNOE, with a correlation of -0.67 and a confidence interval of [-0.68, -0.66]. (b) Line plot showing per-residue hetNOE or ReLU(1 - RMSF predicted) for the protein ACRIIA4, showing agreement between the two. (c) Two structures of ACRIIA4, colored by hetNOE or ReLU(1 - RMSF predicted), showing agreement between the two.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7axvdjjgnvf66wxvnv4lyg52/bafkreidaurrdcvmpbes7hi2phcaw44pxtpf3sf6pew7hjqdhk2dmmxyia4@jpeg)
June 23, 2025 at 8:42 PM
And we think we can! Thanks to excellent data curation efforts from @hkws.bsky.social, we show that RocketSHP-predicted fluctuations correlate well with experimental hetNOE measurements that capture fast + local movement.
I constantly reference their Supp. Fig. 3 when I’m trying to describe the method to people— maybe not exactly what you’re looking for but imo extremely informative.

April 5, 2025 at 8:30 AM
I constantly reference their Supp. Fig. 3 when I’m trying to describe the method to people— maybe not exactly what you’re looking for but imo extremely informative.
Finally, we show how users can use MINT through two case studies:
MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.
MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.


March 12, 2025 at 5:37 PM
Finally, we show how users can use MINT through two case studies:
MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.
MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.
We show MINT works for diverse and challenging interaction tasks!
It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.
It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.

March 12, 2025 at 5:37 PM
We show MINT works for diverse and challenging interaction tasks!
It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.
It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.
MINT sets new benchmarks!
It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈
It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈

March 12, 2025 at 5:37 PM
MINT sets new benchmarks!
It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈
It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈
MINT is built on ESM-2 but adds a cross-chain attention mechanism to preserve inter-sequence relationships.
We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.
We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.

March 12, 2025 at 5:37 PM
MINT is built on ESM-2 but adds a cross-chain attention mechanism to preserve inter-sequence relationships.
We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.
We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.
Traditional PLMs struggle with PPIs since they model proteins independently. Previous approaches concatenated embeddings or sequences—leading to lost inter-residue context. We fix this with 🌿 MINT, which allows multiple interacting sequences as input.

March 12, 2025 at 5:37 PM
Traditional PLMs struggle with PPIs since they model proteins independently. Previous approaches concatenated embeddings or sequences—leading to lost inter-residue context. We fix this with 🌿 MINT, which allows multiple interacting sequences as input.
In my personal favorite figure, we annotate a diverse set of ~3.5 million sequences across kingdoms to see the distribution of symmetry use across life -- another example of the cool things you can do with PLM fine-tuning + lightweight classification + genome scale inference!

March 3, 2025 at 4:43 PM
In my personal favorite figure, we annotate a diverse set of ~3.5 million sequences across kingdoms to see the distribution of symmetry use across life -- another example of the cool things you can do with PLM fine-tuning + lightweight classification + genome scale inference!
I'm at @biophysicalsoc.bsky.social #BPS2025 with a bunch of folks from the Structural and Molecular Biophysics @flatironinstitute.org group-- come say hi and check out our posters/talks! @sonyahanson.bsky.social @pilarcossio.bsky.social @miroastore.bsky.social
February 15, 2025 at 9:54 PM
I'm at @biophysicalsoc.bsky.social #BPS2025 with a bunch of folks from the Structural and Molecular Biophysics @flatironinstitute.org group-- come say hi and check out our posters/talks! @sonyahanson.bsky.social @pilarcossio.bsky.social @miroastore.bsky.social
If you're at @neuripsconf.bsky.social @workshopmlsb.bsky.social tomorrow, make sure you stop by our poster presenting a paired-protein language model for modeling protein-protein interactions. Varun did a great job spearheading this work! #NeurIPS2024

December 14, 2024 at 7:10 PM
If you're at @neuripsconf.bsky.social @workshopmlsb.bsky.social tomorrow, make sure you stop by our poster presenting a paired-protein language model for modeling protein-protein interactions. Varun did a great job spearheading this work! #NeurIPS2024

