
Gina El Nesr
@ginaelnesr.bsky.social
stanford phd • nsf grf • jhu bs • deep learning for protein design & dynamics • oly weightlifter 🏋🏽♀️• 🇪🇬
gelnesr.github.io
gelnesr.github.io
Pinned
Gina El Nesr
@ginaelnesr.bsky.social
· Mar 20
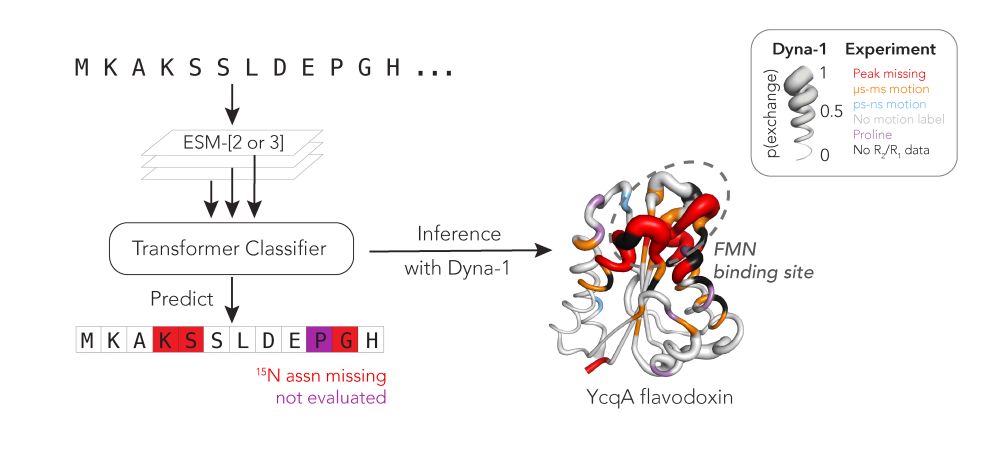
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics?
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
MLSB is coming to Copenhagen, DK 🎉
Submission is under the same portal, just simply indicate your location preference(s).
Submission is under the same portal, just simply indicate your location preference(s).
You asked and we listened... @workshopmlsb.bsky.social is excited to be expanding to Copenhagen, DK at @euripsconf.bsky.social 🎉
Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫
Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫

September 12, 2025 at 12:45 PM
MLSB is coming to Copenhagen, DK 🎉
Submission is under the same portal, just simply indicate your location preference(s).
Submission is under the same portal, just simply indicate your location preference(s).
Can't find the words so heres a diagram, apologies for bad handwriting. Higher energy states that are in between the two conformations you care about are both more combinatorially plentiful, and rarer, and only a few of them matter. mAb CDRs can interconvert on 10s of µs so it matters for those too

August 26, 2025 at 2:14 AM
Reposted by Gina El Nesr
... each independent simulation is 500 ns long, almost certainly too short to sample the sorts of large-scale functionally relevant conformational changes that ML methods should be learning from, which means I must once again repost this excerpt from Janson et al


August 21, 2025 at 1:41 PM
... each independent simulation is 500 ns long, almost certainly too short to sample the sorts of large-scale functionally relevant conformational changes that ML methods should be learning from, which means I must once again repost this excerpt from Janson et al
Reposted by Gina El Nesr
Not sure how many scientists here have tried Claude Code or similar command line coding assistants. I had a complicated family property tax problem that was best solved by a brute force Monte Carlo simulation approach, so I spent a few days coding up and analyzing a model with Claude Code.
August 12, 2025 at 12:47 PM
Not sure how many scientists here have tried Claude Code or similar command line coding assistants. I had a complicated family property tax problem that was best solved by a brute force Monte Carlo simulation approach, so I spent a few days coding up and analyzing a model with Claude Code.
Reposted by Gina El Nesr
I am super excited to announce that I will be starting my lab at the Department of Pharmacology of the University of Zurich in Switzerland next year!
July 25, 2025 at 7:48 AM
I am super excited to announce that I will be starting my lab at the Department of Pharmacology of the University of Zurich in Switzerland next year!
The MLSB workshop will be in San Diego, CA (co-located with NeurIPS) this year for its 6th edition in December 🧬🔬
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟

July 28, 2025 at 3:40 PM
The MLSB workshop will be in San Diego, CA (co-located with NeurIPS) this year for its 6th edition in December 🧬🔬
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟
molecular cat-formations
July 13, 2025 at 3:40 AM
molecular cat-formations
Enzyme design! Benchmark data! Wet-lab validation! Expression & activity assays!! ✨
1/4
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience

July 8, 2025 at 5:04 PM
Enzyme design! Benchmark data! Wet-lab validation! Expression & activity assays!! ✨
Reposted by Gina El Nesr
1/4
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience

July 8, 2025 at 4:26 PM
1/4
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
Reposted by Gina El Nesr
Noooo😭
This was the BEST workshop at NeurIPS!!
This was the BEST workshop at NeurIPS!!
Unfortunately, the MLSB Workshop @ NeurIPS (@workshopmlsb.bsky.social) was rejected this year.
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…

July 7, 2025 at 5:37 PM
Noooo😭
This was the BEST workshop at NeurIPS!!
This was the BEST workshop at NeurIPS!!
Reposted by Gina El Nesr
Folddisco finds similar (dis)continuous 3D motifs in large protein structure databases. Its efficient index enables fast uncharacterized active site annotation, protein conformational state analysis and PPI interface comparison. 1/9🧶🧬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
July 7, 2025 at 8:21 AM
Folddisco finds similar (dis)continuous 3D motifs in large protein structure databases. Its efficient index enables fast uncharacterized active site annotation, protein conformational state analysis and PPI interface comparison. 1/9🧶🧬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
Unfortunately, the MLSB Workshop @ NeurIPS (@workshopmlsb.bsky.social) was rejected this year.
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…

July 7, 2025 at 2:21 PM
Unfortunately, the MLSB Workshop @ NeurIPS (@workshopmlsb.bsky.social) was rejected this year.
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…
Reposted by Gina El Nesr
Speak up to prevent a major loss for the field. CASP, which helped shaping structural biology including AlphaFold is on the verge of shutting down. NIH funding has lapsed, UC Davis support ends and the core team is being let go. Painfully shortsighted.
Exclusive: Famed protein structure competition nears end as NIH grant money runs out
Agency silent on funding renewal for contest that inspired creation of AIs that predicted how proteins would fold
www.science.org
July 4, 2025 at 12:07 PM
Speak up to prevent a major loss for the field. CASP, which helped shaping structural biology including AlphaFold is on the verge of shutting down. NIH funding has lapsed, UC Davis support ends and the core team is being let go. Painfully shortsighted.
Reposted by Gina El Nesr
Exclusive: An international scientific competition widely credited with spurring the development of artificial intelligence for biology appears to be on its deathbed. scim.ag/44ukS90
Exclusive: Famed protein structure competition nears end as NIH grant money runs out
Agency silent on funding renewal for contest that inspired creation of AIs that predicted how proteins would fold
scim.ag
July 2, 2025 at 10:00 PM
Exclusive: An international scientific competition widely credited with spurring the development of artificial intelligence for biology appears to be on its deathbed. scim.ag/44ukS90
Reposted by Gina El Nesr
The dependence of the amino acid backbone conformation on the translated synonymous codon is not statistically significant
doi.org/10.1073/pnas...
[Following: Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon doi.org/10.1038/s414...
doi.org/10.1073/pnas...
[Following: Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon doi.org/10.1038/s414...
The dependence of the amino acid backbone conformation on the translated synonymous codon is not statistically significant | PNAS
The correlation between synonymous codon usage and secondary structure in translated
proteins has been widely demonstrated. This usage plays a capi...
doi.org
July 2, 2025 at 3:27 PM
The dependence of the amino acid backbone conformation on the translated synonymous codon is not statistically significant
doi.org/10.1073/pnas...
[Following: Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon doi.org/10.1038/s414...
doi.org/10.1073/pnas...
[Following: Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon doi.org/10.1038/s414...
Ran some sequence design with LigandMPNN and it... designed a protein with an (almost) His-tag 🤔

June 23, 2025 at 6:42 PM
Ran some sequence design with LigandMPNN and it... designed a protein with an (almost) His-tag 🤔
someone said we need a meme

May 12, 2025 at 3:35 PM
someone said we need a meme
Reposted by Gina El Nesr
"De novo prediction of protein structural dynamics"
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
delalamo.xyz
April 27, 2025 at 2:16 PM
"De novo prediction of protein structural dynamics"
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
Reposted by Gina El Nesr
While this paper looks interesting, let me just say (again) that (essentially all) NMR ensembles in the PDB are NOT thermodynamic ensembles or meant to represent these. They are "uncertainty ensembles" and using them to benchmark machine learning (or other) models of dynamics is not a good idea.

May 4, 2025 at 2:58 PM
While this paper looks interesting, let me just say (again) that (essentially all) NMR ensembles in the PDB are NOT thermodynamic ensembles or meant to represent these. They are "uncertainty ensembles" and using them to benchmark machine learning (or other) models of dynamics is not a good idea.
Reposted by Gina El Nesr
If we wanna make accurate boltzmann samplers, we gotta know what distributions they should be sampling 😁 These change in the presence/absence of ligands, and there are multiple systems for which this is well-understood by this point experimentally.
April 28, 2025 at 12:59 PM
If we wanna make accurate boltzmann samplers, we gotta know what distributions they should be sampling 😁 These change in the presence/absence of ligands, and there are multiple systems for which this is well-understood by this point experimentally.
Reposted by Gina El Nesr
Really nice resource from @delalamo.xyz !!
Wanna mention the "AdK test" we're noticing newer DL methods fall short on:
many proteins (like AdK) that have apo/holo conf change sample both the apo/holo state even without ligand bound. But w/o ligand bound, they *are mainly in the apo state*
Wanna mention the "AdK test" we're noticing newer DL methods fall short on:
many proteins (like AdK) that have apo/holo conf change sample both the apo/holo state even without ligand bound. But w/o ligand bound, they *are mainly in the apo state*

April 28, 2025 at 12:59 PM
Really nice resource from @delalamo.xyz !!
Wanna mention the "AdK test" we're noticing newer DL methods fall short on:
many proteins (like AdK) that have apo/holo conf change sample both the apo/holo state even without ligand bound. But w/o ligand bound, they *are mainly in the apo state*
Wanna mention the "AdK test" we're noticing newer DL methods fall short on:
many proteins (like AdK) that have apo/holo conf change sample both the apo/holo state even without ligand bound. But w/o ligand bound, they *are mainly in the apo state*
Super excited to be presenting Dyna-1 next week with @hkws.bsky.social !!
Next Tues (4/29) at **4:30PM** ET, we will have @ginaelnesr.bsky.social @hkws.bsky.social present "Learning millisecond protein dynamics from what is missing in NMR spectra"
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Learning millisecond protein dynamics from what is missing in NMR spectra
Many proteins’ biological functions rely on interconversions between multiple conformations occurring at micro-to millisecond (µs-ms) timescales. A lack of standardized, large-scale experimental data ...
biorxiv.org
April 22, 2025 at 9:40 PM
Super excited to be presenting Dyna-1 next week with @hkws.bsky.social !!
Reposted by Gina El Nesr
Next Tues (4/29) at **4:30PM** ET, we will have @ginaelnesr.bsky.social @hkws.bsky.social present "Learning millisecond protein dynamics from what is missing in NMR spectra"
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Learning millisecond protein dynamics from what is missing in NMR spectra
Many proteins’ biological functions rely on interconversions between multiple conformations occurring at micro-to millisecond (µs-ms) timescales. A lack of standardized, large-scale experimental data ...
biorxiv.org
April 22, 2025 at 9:08 PM
Next Tues (4/29) at **4:30PM** ET, we will have @ginaelnesr.bsky.social @hkws.bsky.social present "Learning millisecond protein dynamics from what is missing in NMR spectra"
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Reposted by Gina El Nesr
Any NN trained on publicly available MD data needs this disclaimer: trajs that are 100s of ns long are heavily influenced by the starting state and are unlikely to teach the kinds of trends in equilibrium dynamics that scientists are interested in predicting
A great point by the authors about whether short MD simulations from mdCATH & other sources are sufficient as training data for NNs if the goal is to learn the full weighted distribution of the conformational ensemble. They think it isn't!
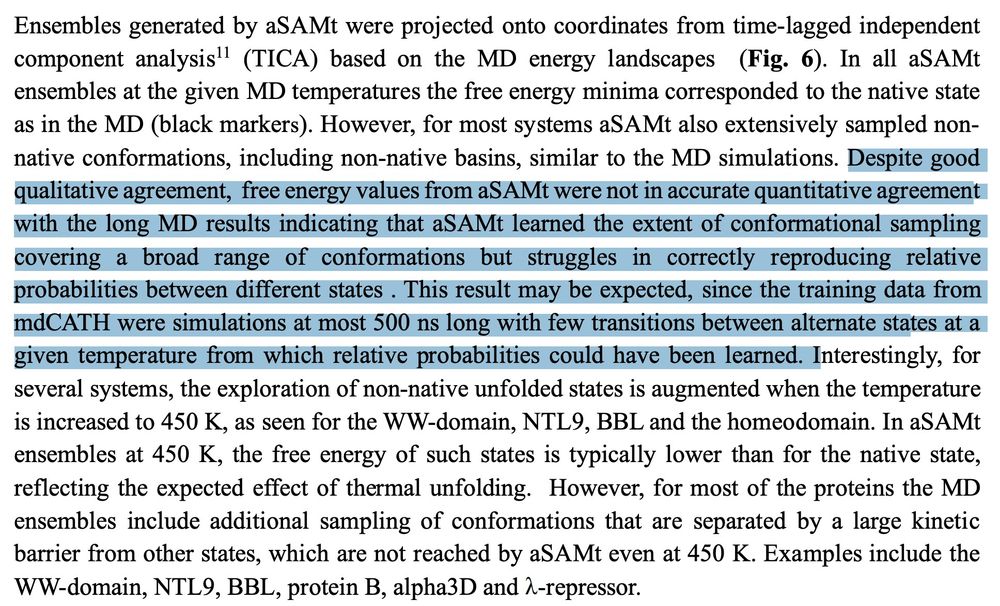
April 21, 2025 at 7:40 AM
Any NN trained on publicly available MD data needs this disclaimer: trajs that are 100s of ns long are heavily influenced by the starting state and are unlikely to teach the kinds of trends in equilibrium dynamics that scientists are interested in predicting

