present "Deep learning guided design of protease substrates"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!

present "Deep learning guided design of protease substrates"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Paper: www.biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: www.science.org/doi/10.1126/...
Sign up on our website for zoom links!

Paper: www.science.org/doi/10.1126/...
Sign up on our website for zoom links!
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!
Paper: www.nature.com/articles/s41...
Sign up on our website for zoom links!
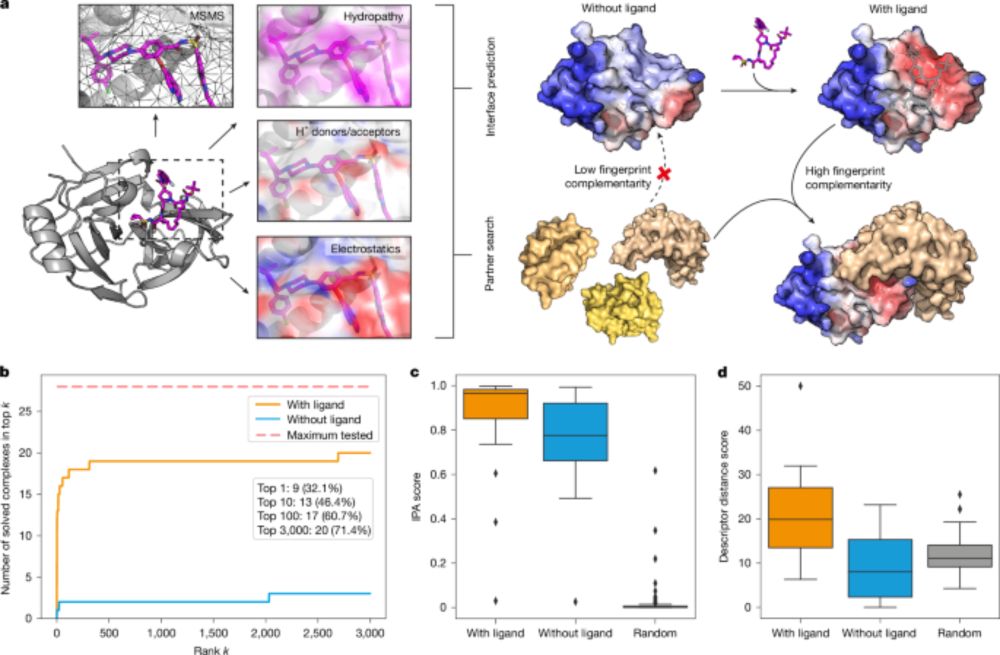
Paper: www.nature.com/articles/s41...
Sign up on our website for zoom links!
present "Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences"
Paper: arxiv.org/abs/2412.07763
Sign up on our website for zoom links!

present "Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences"
Paper: arxiv.org/abs/2412.07763
Sign up on our website for zoom links!
Paper: openreview.net/forum?id=TVQ...
Sign up on our website for zoom links!

Paper: openreview.net/forum?id=TVQ...
Sign up on our website for zoom links!
Paper: openreview.net/forum?id=0ct...
Sign up on our website for zoom links!

Paper: openreview.net/forum?id=0ct...
Sign up on our website for zoom links!
Paper: arxiv.org/abs/2411.06090
Sign up on our website to receive Zoom links!

Paper: arxiv.org/abs/2411.06090
Sign up on our website to receive Zoom links!
will present "InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!

will present "InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders"
Paper: www.biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!
Seyone Chithrananda will present "Mapping the combinatorial coding between olfactory receptors and perception with deep learning"
Paper: biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!

Seyone Chithrananda will present "Mapping the combinatorial coding between olfactory receptors and perception with deep learning"
Paper: biorxiv.org/content/10.1...
Sign up on our website to receive Zoom links!

