
gelnesr.github.io
Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟

Stay tuned @workshopmlsb.bsky.social as we share details about the stellar lineup of speakers, the official call for papers, and other announcements!🌟
Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…

Feedback from the deciding committee indicates it was a coin flip decision, with 283 proposals & a number related to “computational biology”
More on the future of MLSB soon…


Here are some examples comparing Dyna-1 predictions with NMR experimental data, including Olfactory marker protein, Protein Tyrosine-phosphatase A, and BPTI (and BPTI’s ms-timescale MD traj).
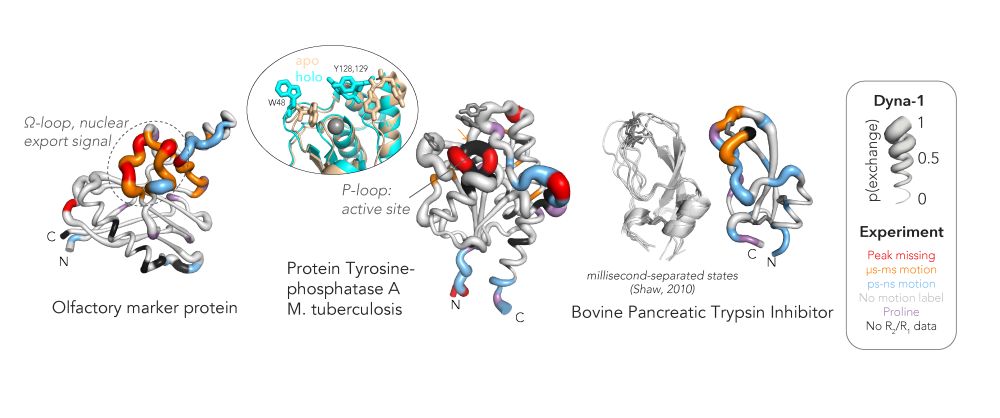
Here are some examples comparing Dyna-1 predictions with NMR experimental data, including Olfactory marker protein, Protein Tyrosine-phosphatase A, and BPTI (and BPTI’s ms-timescale MD traj).
Notably, this model can take in a sequence, structure, or sequence & structure as input. While Dyna-1 performance is best using both, using just one as input still does well.

Notably, this model can take in a sequence, structure, or sequence & structure as input. While Dyna-1 performance is best using both, using just one as input still does well.
So, I did some sweeps…

So, I did some sweeps…
To start, 45% of the proteins had ≥1 NMR structure but no X-ray/EM structures
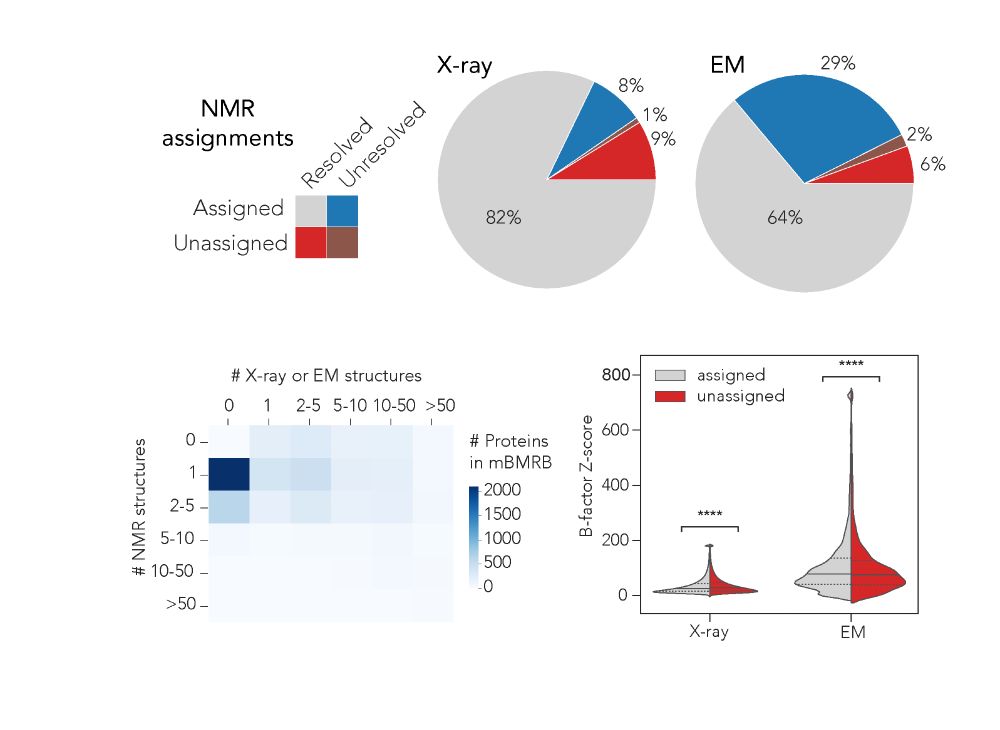
To start, 45% of the proteins had ≥1 NMR structure but no X-ray/EM structures
Where do we find these chemical shift datasets? The BMRB!
Hannah curated another labeled dataset: the "mBMRB"
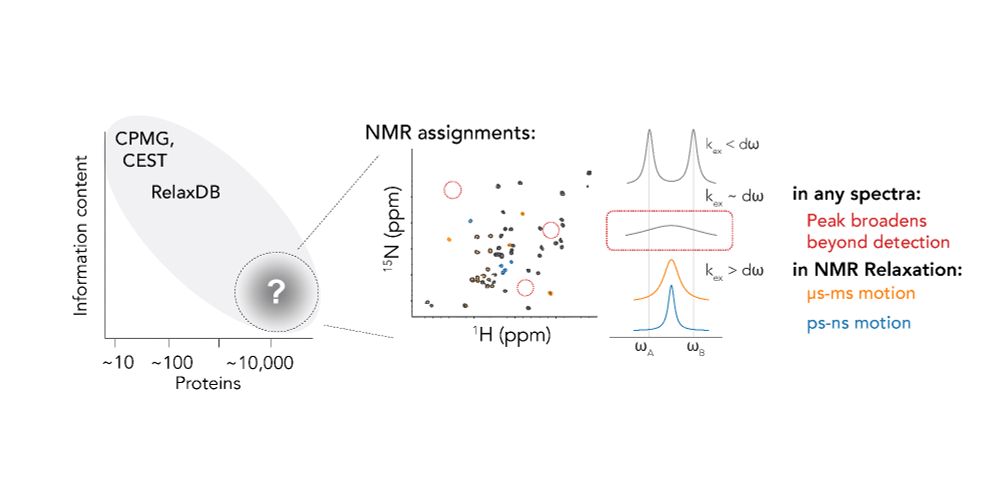
Where do we find these chemical shift datasets? The BMRB!
Hannah curated another labeled dataset: the "mBMRB"

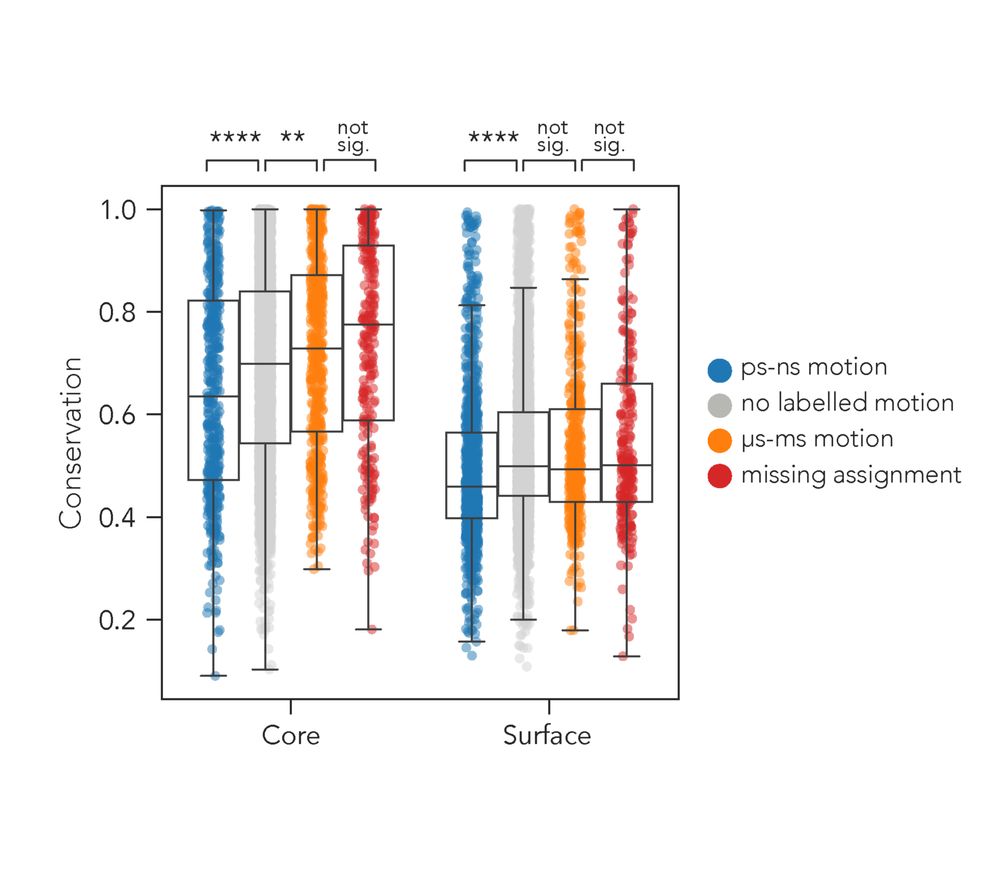
That's Rex, a measurable from NMR. Rex increases when an atom moves at the µs-ms timescale.
There is no DB for that, so Hannah curated "RelaxDB"
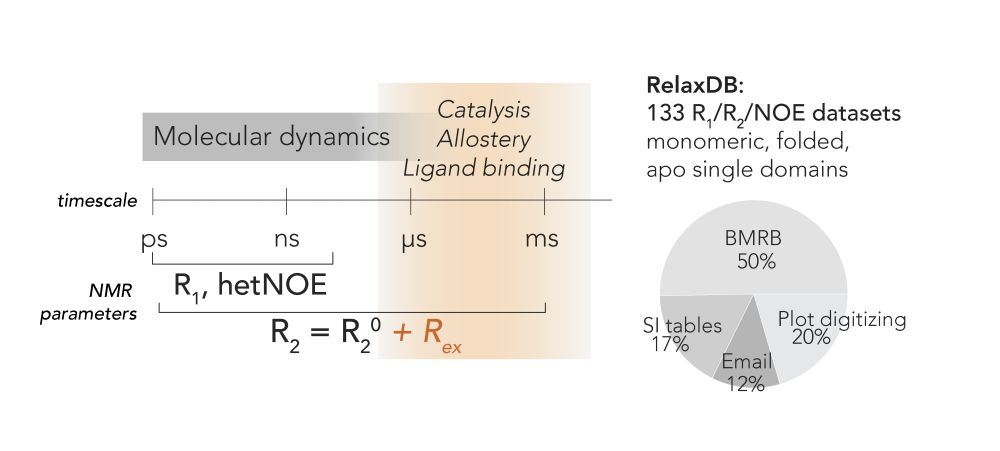
That's Rex, a measurable from NMR. Rex increases when an atom moves at the µs-ms timescale.
There is no DB for that, so Hannah curated "RelaxDB"
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
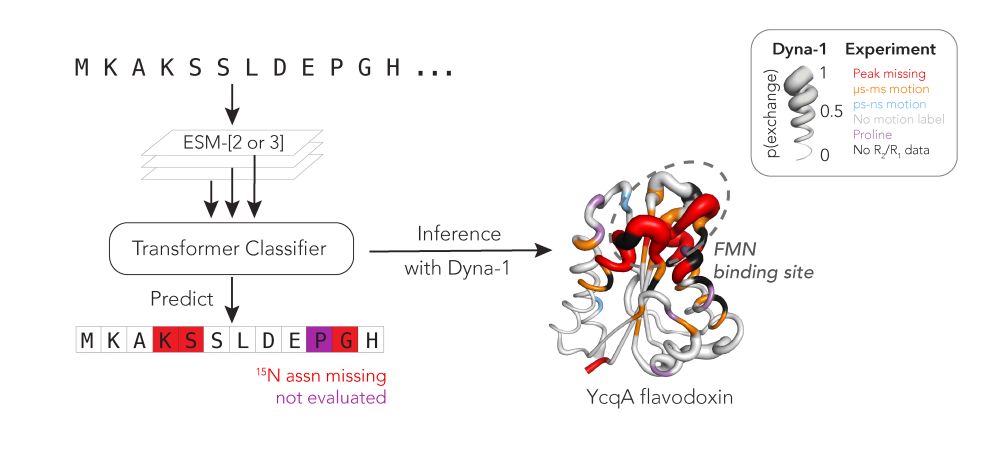
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵


@workshopmlsb.bsky.social


@workshopmlsb.bsky.social

