Maksim Kalutskii
@maksimkalutskii.bsky.social
PhD student at MPINAT. Microtubules and coarse grained simulations.
Pinned
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
Reposted by Maksim Kalutskii
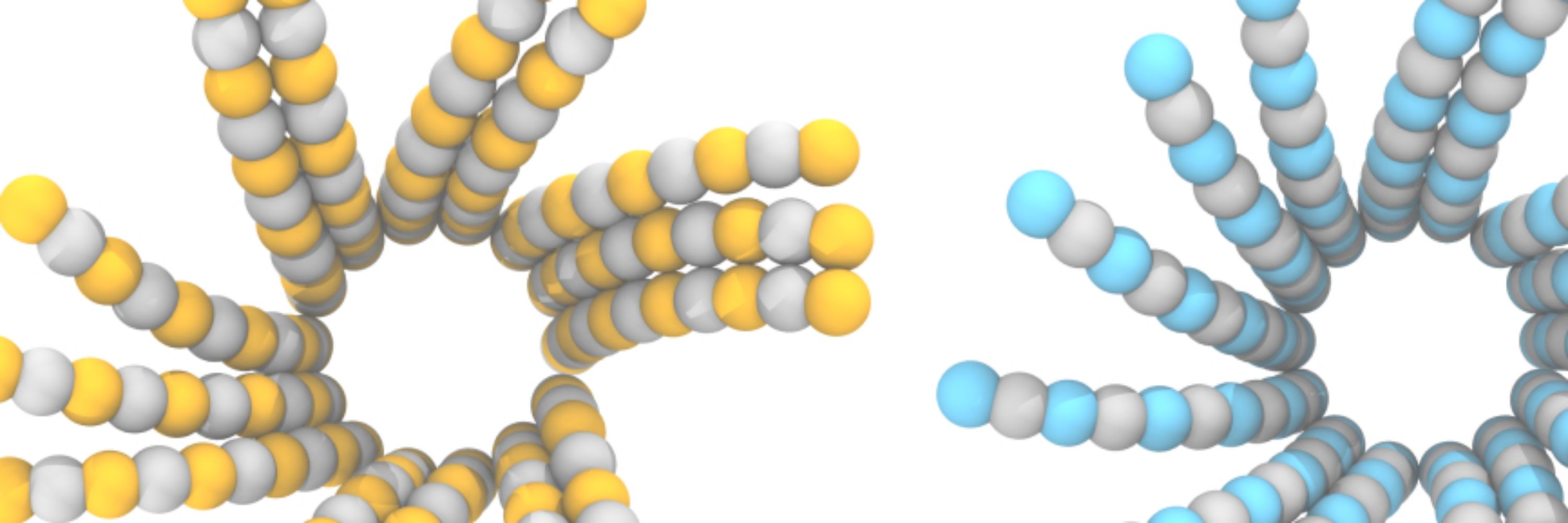
NEAT-DNA: A Chemically Accurate, Sequence-Dependent Coarse-Grained Model for Large-Scale DNA Simulations https://www.biorxiv.org/content/10.1101/2025.11.07.687145v1
November 9, 2025 at 4:47 AM
NEAT-DNA: A Chemically Accurate, Sequence-Dependent Coarse-Grained Model for Large-Scale DNA Simulations https://www.biorxiv.org/content/10.1101/2025.11.07.687145v1
Reposted by Maksim Kalutskii
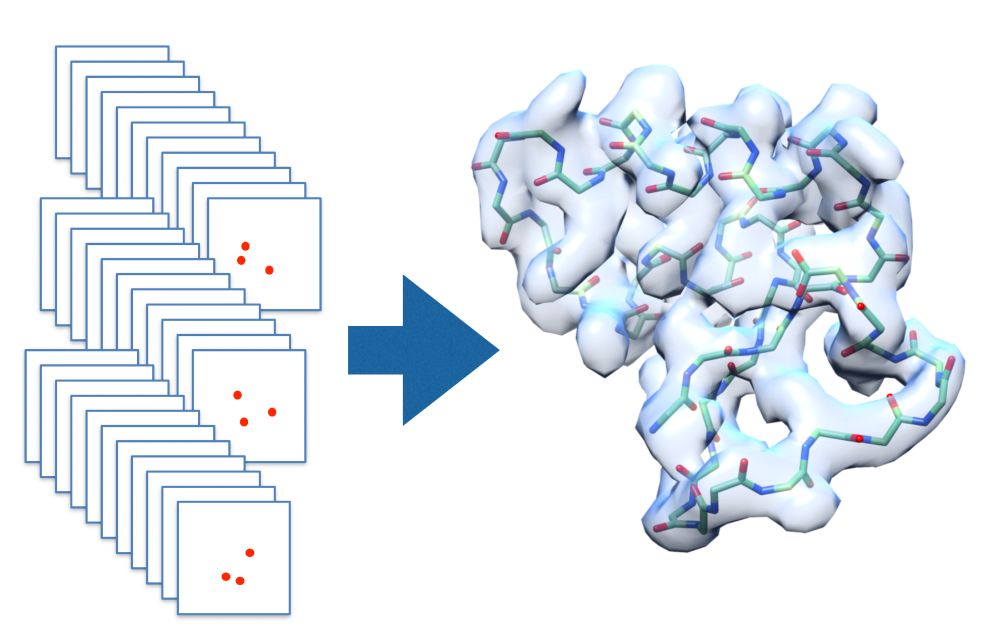
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
CECAM - Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET data
www.cecam.org
November 1, 2025 at 9:47 AM
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
Reposted by Maksim Kalutskii
Are you looking for a PhD opportunity? Or would you like to start your postdoc after having earned your doctorate?
Our Department of Theoretical and Computational Biophysics @compbiophys.bsky.social is hiring: ⬇️
Our Department of Theoretical and Computational Biophysics @compbiophys.bsky.social is hiring: ⬇️
🧐 Are you a computational physicist and expert in probability theory? 👉 Contribute to our project "Theory and Algorithms for Structure Determination from Single Molecule X-Ray Scattering Images" as PhD student or Postdoc! 🔗 Check our job advert for more details www.mpinat.mpg.de/4988122/09-2...
October 29, 2025 at 9:05 AM
Are you looking for a PhD opportunity? Or would you like to start your postdoc after having earned your doctorate?
Our Department of Theoretical and Computational Biophysics @compbiophys.bsky.social is hiring: ⬇️
Our Department of Theoretical and Computational Biophysics @compbiophys.bsky.social is hiring: ⬇️
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
October 27, 2025 at 7:54 AM
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
Reposted by Maksim Kalutskii
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
October 22, 2025 at 2:55 PM
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
Reposted by Maksim Kalutskii
Check out our new work, led by talented @janhuemar.bsky.social, where we uncover that pioneer factor Oct4 remodels chromatin for DNA access not by opening it but by exploiting nucleosome breathing and forming clusters 🔥🔥🔥
🚨 🚨 🚨 New preprint alert!!! 🚨 🚨 🚨
In the past, we have learnt that Oct4 can induce nucleosome breathing on the mono-nucleosome level.
But what happens when you have a fibre of multiple nucleosomes?
www.biorxiv.org/content/10.1...
@rcollepardo.bsky.social @juliamaristany.bsky.social
In the past, we have learnt that Oct4 can induce nucleosome breathing on the mono-nucleosome level.
But what happens when you have a fibre of multiple nucleosomes?
www.biorxiv.org/content/10.1...
@rcollepardo.bsky.social @juliamaristany.bsky.social

October 22, 2025 at 5:10 AM
Check out our new work, led by talented @janhuemar.bsky.social, where we uncover that pioneer factor Oct4 remodels chromatin for DNA access not by opening it but by exploiting nucleosome breathing and forming clusters 🔥🔥🔥
Reposted by Maksim Kalutskii
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

October 20, 2025 at 11:26 AM
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Reposted by Maksim Kalutskii
Dick is one of the most thoughtful folks in cell biology--this is a must read for anyone interested in microtubules!
Perspective from Richard McIntosh describing the history of research on #microtubule polymerization in terms of the ideas, technologies, and observations that have emerged as countless researchers have studied the dynamics of these essential cytoskeletal polymers. rupress.org/jcb/article/...

October 15, 2025 at 6:23 PM
Dick is one of the most thoughtful folks in cell biology--this is a must read for anyone interested in microtubules!
Reposted by Maksim Kalutskii
Eye of the mantis shrimp. Bali/Amed with @cecclementi.bsky.social

September 7, 2025 at 7:55 AM
Eye of the mantis shrimp. Bali/Amed with @cecclementi.bsky.social
Reposted by Maksim Kalutskii
📢 We are hiring #PhD students and #Postdocs at the
@mpi-nat.bsky.social in
in the Department of Theoretical and Computational #Biophysics in Göttingen 🇩🇪! 🔗 check our current job adverts at www.mpinat.mpg.de/grubmueller and find your project. Submit your #application today! #job
@mpi-nat.bsky.social in
in the Department of Theoretical and Computational #Biophysics in Göttingen 🇩🇪! 🔗 check our current job adverts at www.mpinat.mpg.de/grubmueller and find your project. Submit your #application today! #job

Helmut Grubmüller
www.mpinat.mpg.de
September 2, 2025 at 12:33 PM
📢 We are hiring #PhD students and #Postdocs at the
@mpi-nat.bsky.social in
in the Department of Theoretical and Computational #Biophysics in Göttingen 🇩🇪! 🔗 check our current job adverts at www.mpinat.mpg.de/grubmueller and find your project. Submit your #application today! #job
@mpi-nat.bsky.social in
in the Department of Theoretical and Computational #Biophysics in Göttingen 🇩🇪! 🔗 check our current job adverts at www.mpinat.mpg.de/grubmueller and find your project. Submit your #application today! #job
Reposted by Maksim Kalutskii
👉 Open #PhD and #Postdoc positions in #computational #biophysics @mpi-nat.bsky.social in Göttingen 🇩🇪 in the Computational Biomolecular Dynamics group of Bert deGroot for several projects! Applications welcome! 🔗https://www.mpinat.mpg.de/5088717/24-25?c=645962 📝ausschreibung24-25@mpinat.mpg.de

September 2, 2025 at 12:07 PM
👉 Open #PhD and #Postdoc positions in #computational #biophysics @mpi-nat.bsky.social in Göttingen 🇩🇪 in the Computational Biomolecular Dynamics group of Bert deGroot for several projects! Applications welcome! 🔗https://www.mpinat.mpg.de/5088717/24-25?c=645962 📝ausschreibung24-25@mpinat.mpg.de
Reposted by Maksim Kalutskii
A laser flash melting procedure, followed by rapid revitrification, provides a simple approach to help mitigate the preferred orientation problem that plagues cryo-EM analysis.
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Laser flash melting cryo-EM samples to overcome preferred orientation - Nature Methods
Individual proteins tend to adopt preferred orientations when subjected to vitrification for cryo-electron microscopy analysis. A laser flash melting procedure followed by rapid revitrification provid...
www.nature.com
August 29, 2025 at 6:44 PM
A laser flash melting procedure, followed by rapid revitrification, provides a simple approach to help mitigate the preferred orientation problem that plagues cryo-EM analysis.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Maksim Kalutskii
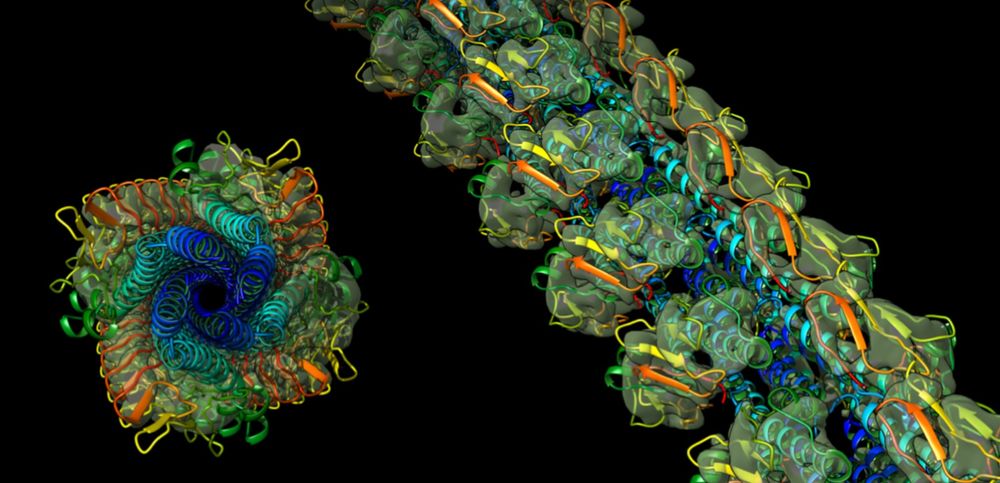
We are recruiting 2 postdocs to work on bacterial motility using a combination of cryoEM, FLM and MD. An exciting collaboration with Kirsty Wan @micromotility.bsky.social Wolfram Moebius @wolframmoebius.bsky.social and Daniel Kattnig. Please see links in the thread below!
August 28, 2025 at 9:13 AM
We are recruiting 2 postdocs to work on bacterial motility using a combination of cryoEM, FLM and MD. An exciting collaboration with Kirsty Wan @micromotility.bsky.social Wolfram Moebius @wolframmoebius.bsky.social and Daniel Kattnig. Please see links in the thread below!
Reposted by Maksim Kalutskii
#PostDoc opportunity in data-driven #SynChem planning at MIT (Cambridge MA USA) duration: 2 years #cheminformatics #DataScience #CompChem #ChemPostDoc #PostDocJobs #chemsky 🧪
coley.mit.edu/positions
coley.mit.edu/positions
Coley Research Group - Positions
Positions
coley.mit.edu
August 12, 2025 at 7:30 AM
#PostDoc opportunity in data-driven #SynChem planning at MIT (Cambridge MA USA) duration: 2 years #cheminformatics #DataScience #CompChem #ChemPostDoc #PostDocJobs #chemsky 🧪
coley.mit.edu/positions
coley.mit.edu/positions
Reposted by Maksim Kalutskii
A few highlights from our new preprint:
On taxol-stabilized microtubules In vitro, wild-type (WT) and phosphor-resistive (AP) tau form well-defined envelopes, while phosphomimetic (E14) tau does not bind cooperatively.
On taxol-stabilized microtubules In vitro, wild-type (WT) and phosphor-resistive (AP) tau form well-defined envelopes, while phosphomimetic (E14) tau does not bind cooperatively.

August 4, 2025 at 5:43 PM
A few highlights from our new preprint:
On taxol-stabilized microtubules In vitro, wild-type (WT) and phosphor-resistive (AP) tau form well-defined envelopes, while phosphomimetic (E14) tau does not bind cooperatively.
On taxol-stabilized microtubules In vitro, wild-type (WT) and phosphor-resistive (AP) tau form well-defined envelopes, while phosphomimetic (E14) tau does not bind cooperatively.
Reposted by Maksim Kalutskii
Nature research paper: Diffusing protein binders to intrinsically disordered proteins
go.nature.com/4lSCdzE
go.nature.com/4lSCdzE
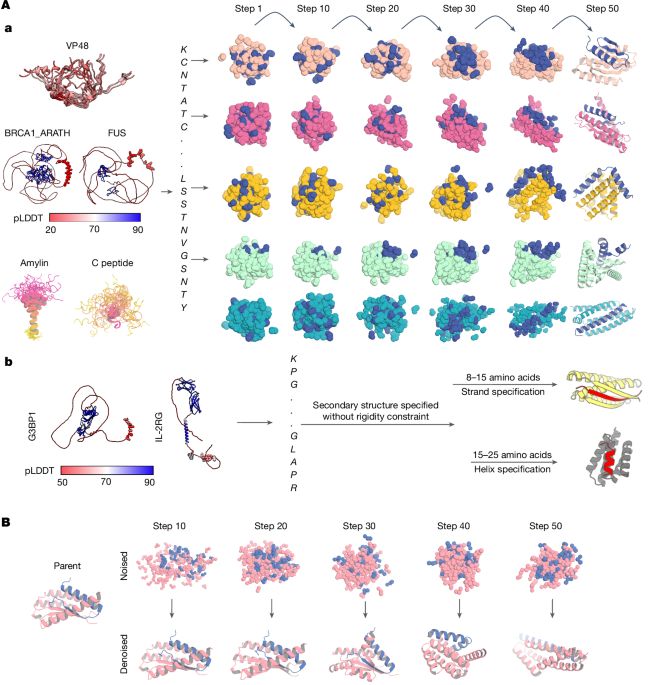
Diffusing protein binders to intrinsically disordered proteins - Nature
Using RFdiffusion, a general method for targeting intrinsically disordered proteins and regions for protein design has been developed.
go.nature.com
July 31, 2025 at 12:58 PM
Nature research paper: Diffusing protein binders to intrinsically disordered proteins
go.nature.com/4lSCdzE
go.nature.com/4lSCdzE
Reposted by Maksim Kalutskii
Intrinsic disorder comes in various flavors ! Using CALVADOS simulations of the Tau protein, we reveal that its disordered dynamics can be interpreted at both residue and domain scales, and that addition of phosphorylations impact both
By @sacquin-mo.bsky.social , Chantal Prévost and I
By @sacquin-mo.bsky.social , Chantal Prévost and I
Across (conformational) space and (relaxation) time: using coarse-grainsimulations to probe the intra- and interdomain dynamics of the Tau protein https://www.biorxiv.org/content/10.1101/2025.07.21.665865v1
July 25, 2025 at 6:59 AM
Intrinsic disorder comes in various flavors ! Using CALVADOS simulations of the Tau protein, we reveal that its disordered dynamics can be interpreted at both residue and domain scales, and that addition of phosphorylations impact both
By @sacquin-mo.bsky.social , Chantal Prévost and I
By @sacquin-mo.bsky.social , Chantal Prévost and I
Reposted by Maksim Kalutskii
Our paper on:
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
July 22, 2025 at 5:58 AM
Our paper on:
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
Reposted by Maksim Kalutskii
Our development of machine-learned transferable coarse-grained models in now on Nat Chem! doi.org/10.1038/s415...
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
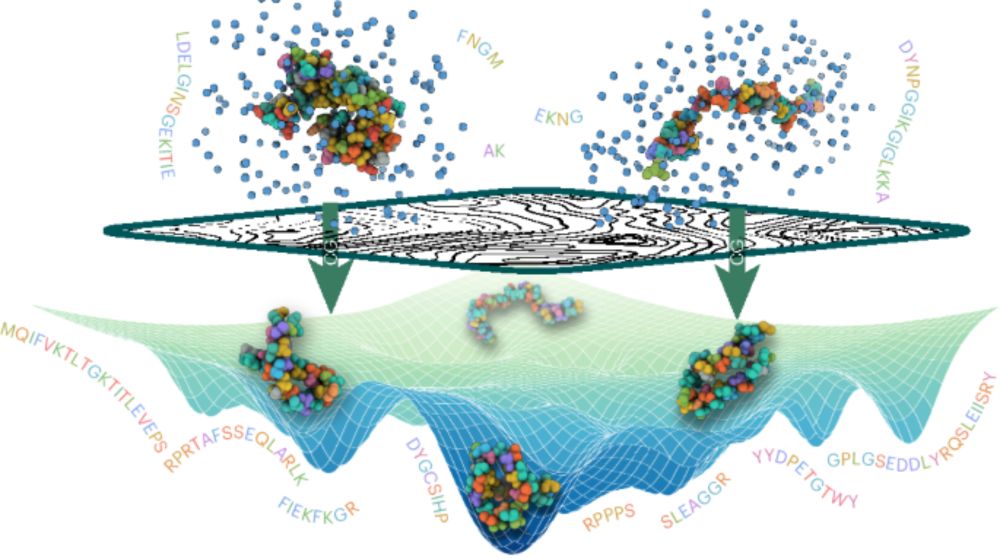
Navigating protein landscapes with a machine-learned transferable coarse-grained model - Nature Chemistry
The development of a universal protein coarse-grained model has been a long-standing challenge. A coarse-grained model with chemical transferability has now been developed by combining deep-learning m...
doi.org
July 18, 2025 at 10:45 AM
Our development of machine-learned transferable coarse-grained models in now on Nat Chem! doi.org/10.1038/s415...
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
Reposted by Maksim Kalutskii
Arriën & Giulio's paper on
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social

July 17, 2025 at 8:54 AM
Arriën & Giulio's paper on
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
Reposted by Maksim Kalutskii
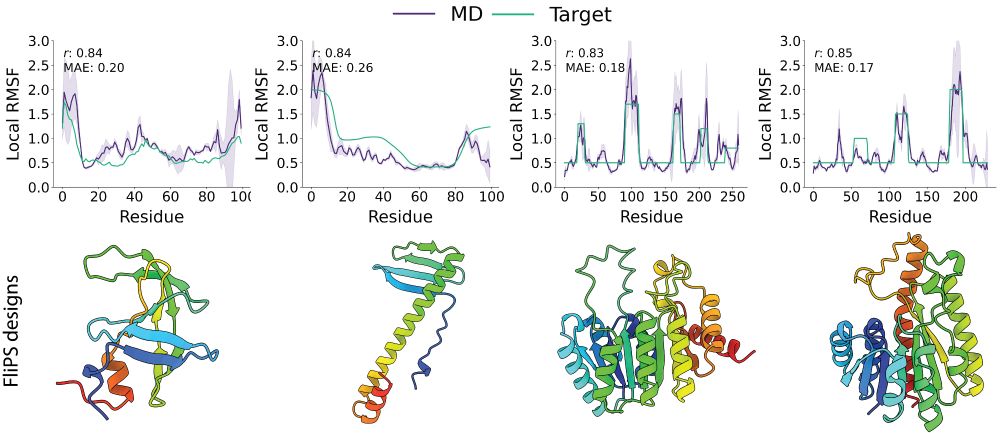
Ever wondered how one can design dynamics into proteins? Check out our work on flexibility conditioned protein structure design at ICML (Poster W-109, Thu. 11am)!
We introduce a flow matching framework for generating protein backbones with custom per-residue flexibility, validated by 300ns of MD.
We introduce a flow matching framework for generating protein backbones with custom per-residue flexibility, validated by 300ns of MD.
1/6 One of the key features of functional proteins is their inherent structural flexibility. In our recent work at #ICML, we introduce flexibility to protein structure design! More in a thread below.
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
July 17, 2025 at 4:32 AM
Ever wondered how one can design dynamics into proteins? Check out our work on flexibility conditioned protein structure design at ICML (Poster W-109, Thu. 11am)!
We introduce a flow matching framework for generating protein backbones with custom per-residue flexibility, validated by 300ns of MD.
We introduce a flow matching framework for generating protein backbones with custom per-residue flexibility, validated by 300ns of MD.
Great work!
New preprint from the lab - the first paper made (almost) entirely here @qmul.bsky.social
We report how human outer kinetochore complexes Ndc80 and Ska form cooperative oligomers, that together stabilise microtubule ends against shortening.
www.biorxiv.org/content/10.1...
Key results below: (1/7)
We report how human outer kinetochore complexes Ndc80 and Ska form cooperative oligomers, that together stabilise microtubule ends against shortening.
www.biorxiv.org/content/10.1...
Key results below: (1/7)
Microtubule end stabilisation by cooperative oligomers of Ska and Ndc80 complexes
During mitosis, properly aligned chromosomes stabilise microtubule ends with the help of kinetochores to ensure timely segregation of chromosomes. Microtubule-binding components of the human outer kinetochore, such as Ndc80 and Ska complexes, are present in multiple copies and together bind several microtubule ends, creating a highly multivalent binding interface. Whereas Ndc80:Ndc80 and Ndc80:microtubule binding is crucial for interface stability, Ndc80 alone in absence of Ska is unable to support stable kinetochore-attachments. Using cryoET, we demonstrate that oligomeric Ndc80:Ska assemblies stabilise microtubule ends against shortening by strengthening lateral contacts between tubulin protofilaments at microtubule plus-ends. We further identify a point mutation within the SKA1 microtubule-binding domain that does not affect microtubule-binding of individual Ska molecules, but does abolish Ska:Ska interactions. Finally, we report that oligomerisation of Ska, in a cooperative fashion together with the Ndc80, is necessary to maintain stable microtubule attachments both in vivo and in vitro. ### Competing Interest Statement The authors have declared no competing interest. BBSRC, BB/X014975/1, BB/W019698/1 Wellcome Trust, https://ror.org/029chgv08, 308895/Z/23/Z
www.biorxiv.org
July 9, 2025 at 6:19 AM
Great work!
Reposted by Maksim Kalutskii
A Minimal Chemo-mechanical Markov Model for Rotary Catalysis of F1-ATPase https://www.biorxiv.org/content/10.1101/2025.06.26.661389v1
June 29, 2025 at 3:48 AM
A Minimal Chemo-mechanical Markov Model for Rotary Catalysis of F1-ATPase https://www.biorxiv.org/content/10.1101/2025.06.26.661389v1
Reposted by Maksim Kalutskii
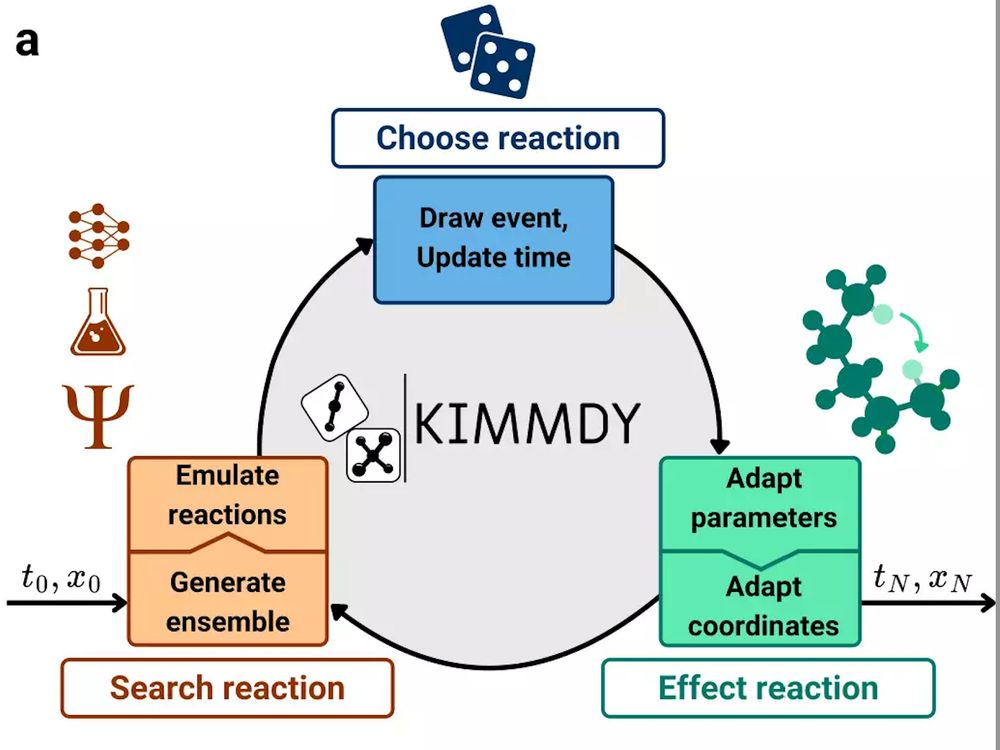
You want to make your simulations reactive, at little cost? Try KIMMDY. Don't hesitate to reach out if you want to include your favorite chemistry. Super happy to see this online, kudos to the team!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
July 7, 2025 at 7:00 AM
You want to make your simulations reactive, at little cost? Try KIMMDY. Don't hesitate to reach out if you want to include your favorite chemistry. Super happy to see this online, kudos to the team!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

