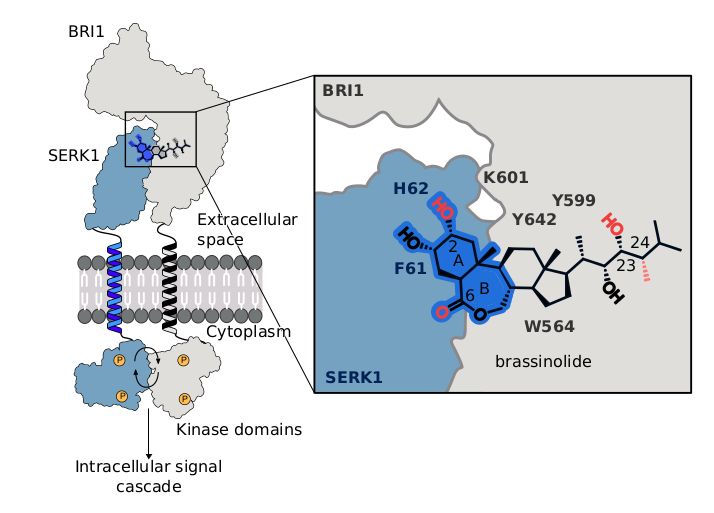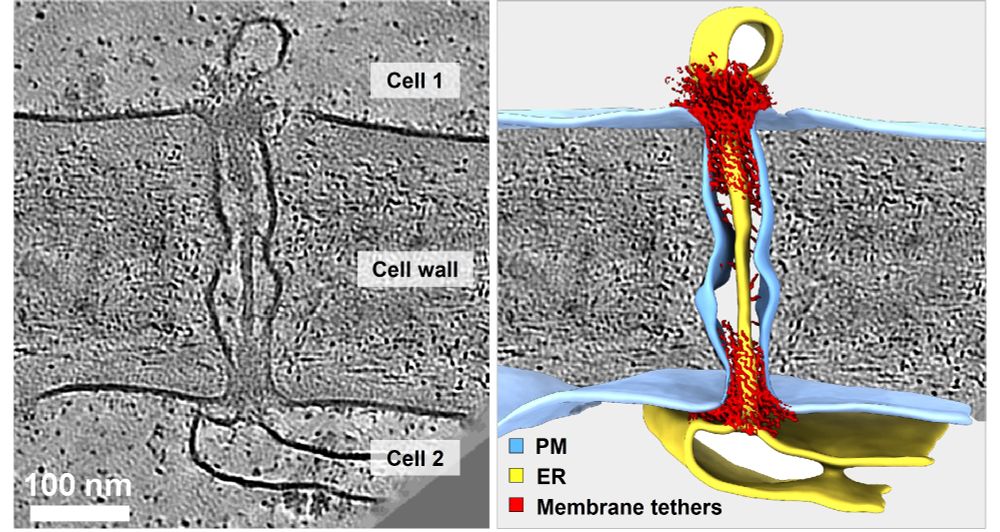
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM
No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...
This dataset + what’s in PDB already would be a decent start.


This dataset + what’s in PDB already would be a decent start.
Please help us by filling out this questionnaire: docs.google.com/forms/d/e/1F...

Please help us by filling out this questionnaire: docs.google.com/forms/d/e/1F...
🧵👇A thread
🧵👇A thread

We asked how can protein complexes diversify without compromising their function and explored this question using the plant #exocyst complex.
www.nature.com/articles/s41...

We asked how can protein complexes diversify without compromising their function and explored this question using the plant #exocyst complex.
www.nature.com/articles/s41...
Advanced imaging techniques and computer modelling to create a detailed structural model of a nuclear pore complex from a seed plant.

Advanced imaging techniques and computer modelling to create a detailed structural model of a nuclear pore complex from a seed plant.
My set for screening (AND):
Side chain heavy atom distance <= 4.2 Å
PAE <= 8 Å
Combined pLDDT >= 140
pDockQ > 0.3
My set for screening (AND):
Side chain heavy atom distance <= 4.2 Å
PAE <= 8 Å
Combined pLDDT >= 140
pDockQ > 0.3


"We can't leave all the fun to the biologists; we should zoom out while they zoom in."
Where to go after AlphaFold? How do we avoid the field becoming a load of half-baked LLMs?
Let me know what you think.
jgreener64.github.io/posts/struct...
"We can't leave all the fun to the biologists; we should zoom out while they zoom in."
@jandevries.bsky.social
@watertoland.bsky.social
www.biorxiv.org/content/10.1...

@jandevries.bsky.social
@watertoland.bsky.social
www.biorxiv.org/content/10.1...
Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...

Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...
This saves some clicking around. Neat!

This saves some clicking around. Neat!
Time to update some sequence alignments 🥲
Apparently, bryophytes time and again include microbial genes in their genomic repertoire, and occupy a langer gene space than vascular plants.
www.nature.com/articles/s41...

Time to update some sequence alignments 🥲
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
🖥️ github.com/NVIDIA-Digit...
🗎 arxiv.org/abs/2507.09466
🖥️ github.com/NVIDIA-Digit...
🗎 arxiv.org/abs/2507.09466
Soon at @spirochrome.com
Led by O.Mercey and @lreymond.bsky.social, in collab with @dudinlab.bsky.social and @marinelap.bsky.social

Soon at @spirochrome.com
Led by O.Mercey and @lreymond.bsky.social, in collab with @dudinlab.bsky.social and @marinelap.bsky.social
#live-imaging
buff.ly/WsOrSlR
#live-imaging