

Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...
Huge collaborative effort! So glad to see the community already using it to develop new resources & tools.
Check it out here: shorturl.at/z4i4c
#CryoEM #CryoET

Huge collaborative effort! So glad to see the community already using it to develop new resources & tools.
Check it out here: shorturl.at/z4i4c
#CryoEM #CryoET
Averaging-free direct visualization of ribosome translational state! #teamtomo #cryoet

Averaging-free direct visualization of ribosome translational state! #teamtomo #cryoet
www.umu.se/en/news/umea...

www.umu.se/en/news/umea...
Henipaviruses, like Nipah and Hendra, package their genomes inside helical shells built by thousands of nucleoproteins. These nucleocapsids are essential to protect the viral RNA, but how do they ever let the polymerase in to read the sequence?
👇

Henipaviruses, like Nipah and Hendra, package their genomes inside helical shells built by thousands of nucleoproteins. These nucleocapsids are essential to protect the viral RNA, but how do they ever let the polymerase in to read the sequence?
👇
Please try, we need feedback 🧪🧶🧬🔬

Please try, we need feedback 🧪🧶🧬🔬
www.biorxiv.org/content/10.1...
#teamtomo

www.biorxiv.org/content/10.1...
#teamtomo
Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...

Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...

Here, we used #CryoET to visualise mitochondrial proteostatic stress and together with SPA #CryoEM shed light into the functional cycle of the Hsp60:10 chaperone system. #TeamTomo
🔗 www.biorxiv.org/content/10.1...
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
www.biozentrum.unibas.ch/open-positio...
#biozentrum #University #Basel #Professor #Structural #Biology #Biophysics #Biological #Imaging

www.biozentrum.unibas.ch/open-positio...
#biozentrum #University #Basel #Professor #Structural #Biology #Biophysics #Biological #Imaging
Link: www.biorxiv.org/content/10.1...

Link: www.biorxiv.org/content/10.1...
buff.ly/j3TSIkn
Contents: weird #NPCs in yeast and flies, #CLEM, in situ #cryoET and more!
Made possible by the amazing @becklab.bsky.social team at @mpibp.bsky.social and collaborators! #TeamTomo

Contents: weird #NPCs in yeast and flies, #CLEM, in situ #cryoET and more!
Made possible by the amazing @becklab.bsky.social team at @mpibp.bsky.social and collaborators! #TeamTomo
A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo

A massive team effort led by @leonieanton.bsky.social Meseret Haile & Wenjing Cheng in collaboration with Jerzy Dziekan & the Alan Cowman! #TeamTomo
Open #PhDposition in structural virology in my group! Join us in Umeå, Sweden to uncover how arboviruses remodel infected cells. In situ #cryoET and #cryoEM combined with #virology, #cellbiology, and #biophysics.
Deadline 7 September. More info: www.carlsonlab.se/join/
Open #PhDposition in structural virology in my group! Join us in Umeå, Sweden to uncover how arboviruses remodel infected cells. In situ #cryoET and #cryoEM combined with #virology, #cellbiology, and #biophysics.
Deadline 7 September. More info: www.carlsonlab.se/join/
www.biorxiv.org/content/10.1...
Congrats🎉 to @vasilgaisin.bsky.social and Corina Hadjicharalambous, who studied contractile injection systems directly in hot spring♨️ bacterial mats🦠
#microsky #teamtomo #Chloroflexota
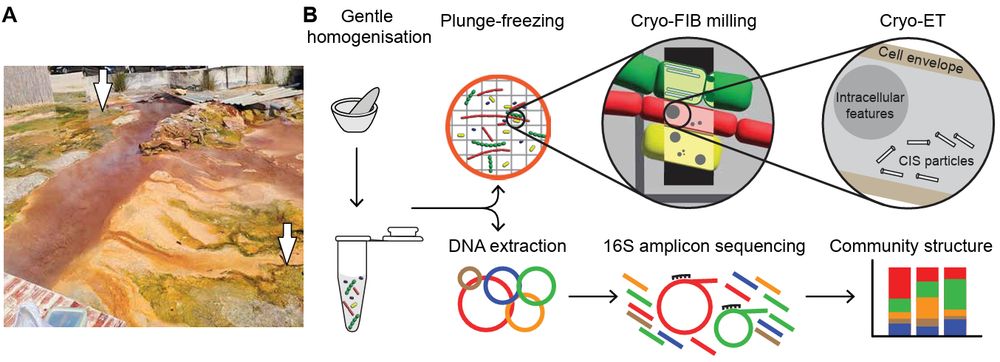
www.biorxiv.org/content/10.1...
Congrats🎉 to @vasilgaisin.bsky.social and Corina Hadjicharalambous, who studied contractile injection systems directly in hot spring♨️ bacterial mats🦠
#microsky #teamtomo #Chloroflexota
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
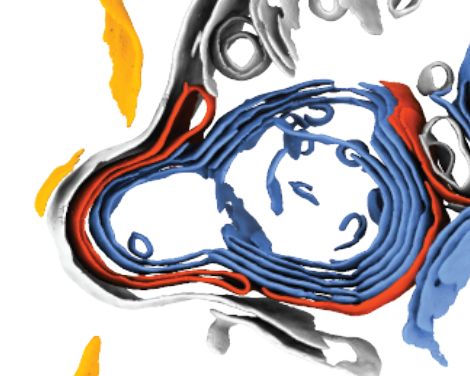
Make my day and check it out!
www.biorxiv.org/content/10.1...
#Cryo-ET #TeamTomo #Cryo-CLEM ❄️🔬❄️🔬❄️🔬❄️🔬
Make my day and check it out!
www.biorxiv.org/content/10.1...
#Cryo-ET #TeamTomo #Cryo-CLEM ❄️🔬❄️🔬❄️🔬❄️🔬
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM
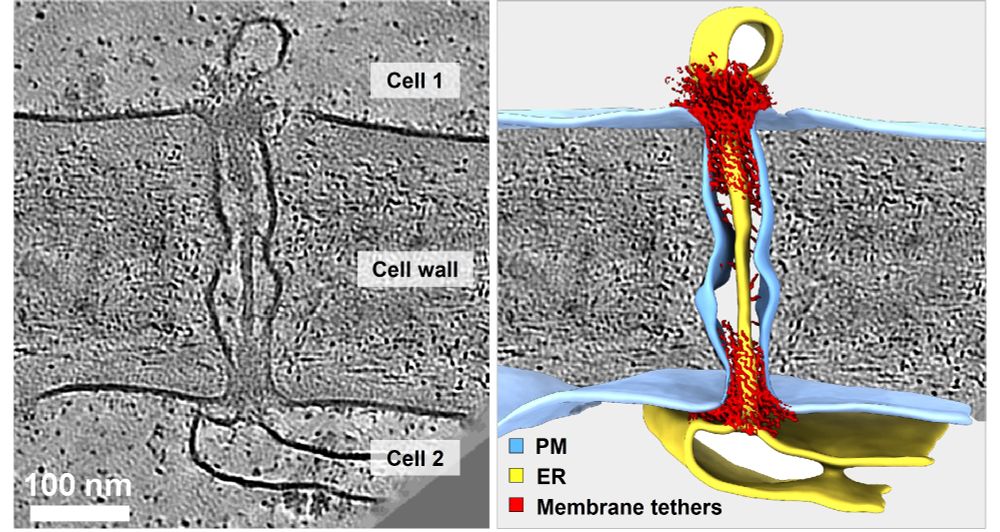
Using #cryoET, #AlphaFold & proteomics, we uncover the native organization of plant intercellular nanopores.
🔗 doi.org/10.1101/2025...
🧪🧵1/n
#teamtomo #PlantScience #Physcomitrium #Arabidopsis #cryoEM
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
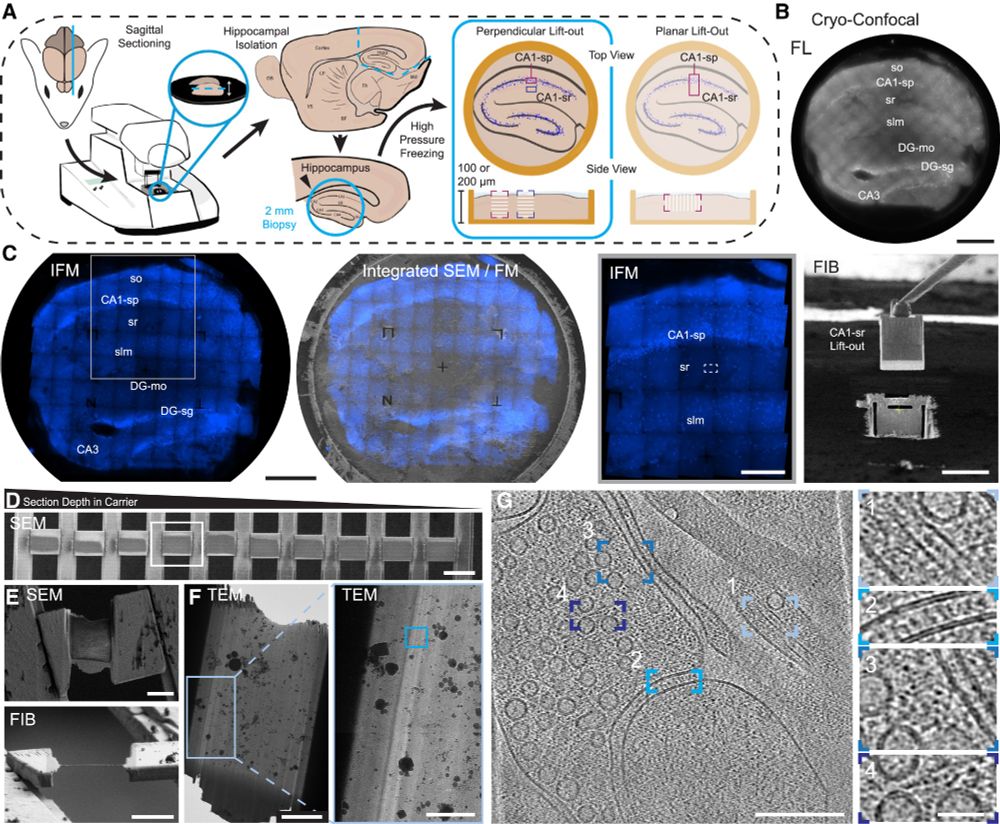
Through the dedication of @glynnca.bsky.social and @cryingem.bsky.social we report a thorough method to image molecular organisation within hippocampus tissue.
Structural biology in tissue is well and truly here!
@rosfrankinst.bsky.social
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...


