

---
#proteomics #prot-paper
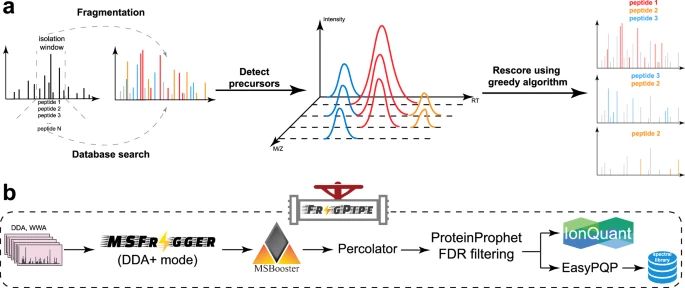
---
#proteomics #prot-paper
pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....

🚀 New Research Paper Alert! 🚀
🦠 Big news for #Bacterial #MassSpec #Proteomics!
2025 has started properly for our team. 🥳

🚀 New Research Paper Alert! 🚀
🦠 Big news for #Bacterial #MassSpec #Proteomics!
2025 has started properly for our team. 🥳
(1) NIH doesn't fund truly high impact research
(2) NIH needs to experiment with alternative approaches to peer review, particularly for high risk-high impact research
1/n
(1) NIH doesn't fund truly high impact research
(2) NIH needs to experiment with alternative approaches to peer review, particularly for high risk-high impact research
1/n
"Bluesky is much better for science. There is much less toxicity, misinformation, and distractions."
www.nature.com/articles/d41...
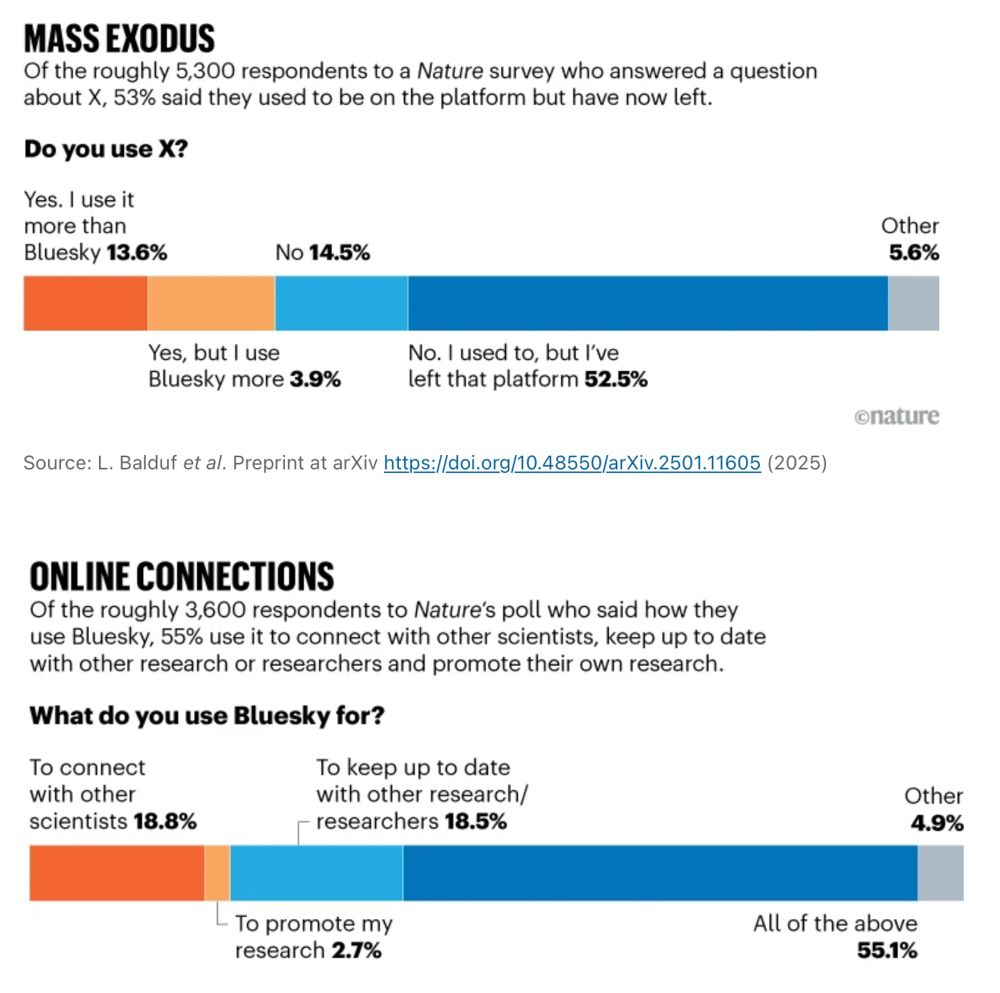
"Bluesky is much better for science. There is much less toxicity, misinformation, and distractions."
www.nature.com/articles/d41...
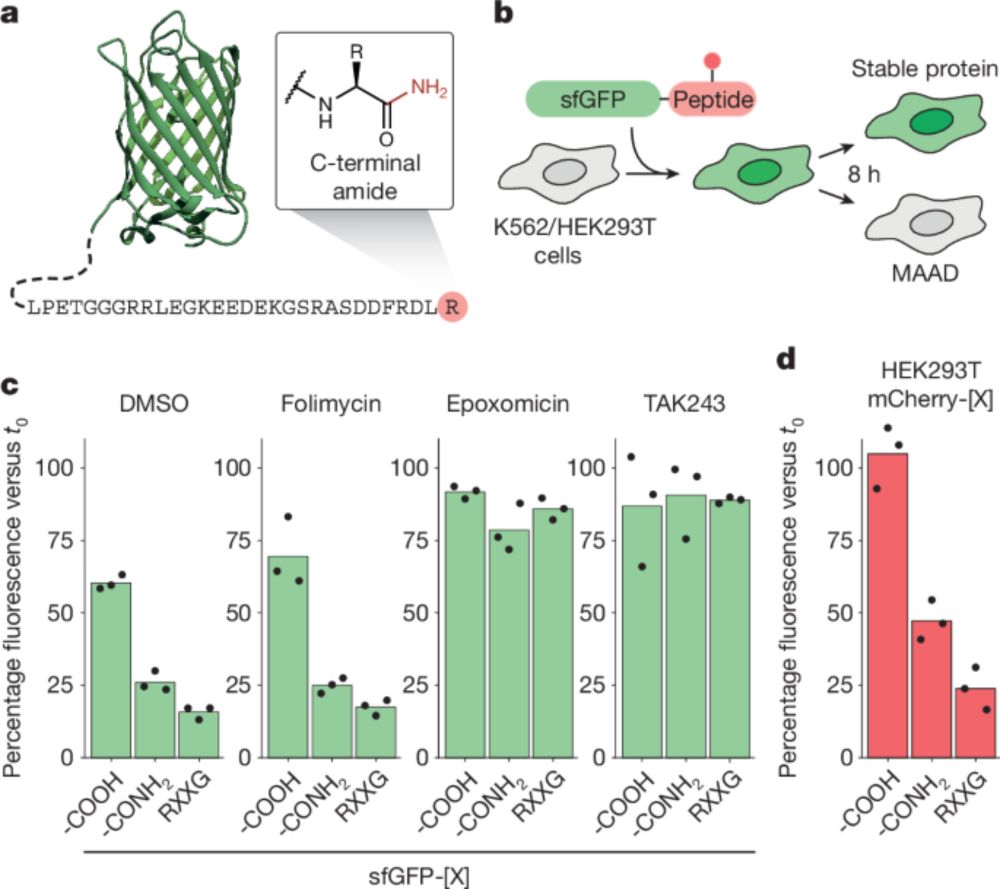
#Acetylation #Methylation #Myristoylation #Ubiquitylation #Oxidation #Arinylation
authors.elsevier.com/sd/article/S...

#Acetylation #Methylation #Myristoylation #Ubiquitylation #Oxidation #Arinylation
authors.elsevier.com/sd/article/S...
www.nature.com/articles/s42...

www.nature.com/articles/s42...
Also the Bluesky proteomics migration is almost complete!
Also the Bluesky proteomics migration is almost complete!
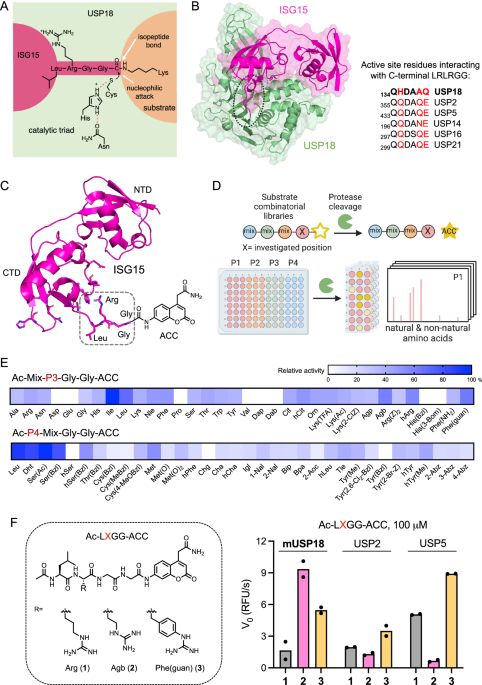
A huge shoutout to my amazing co-authors for their incredible contributions!
Key highlights in replies (1/n)

A huge shoutout to my amazing co-authors for their incredible contributions!
Key highlights in replies (1/n)
Source: Journal of Proteomics
Author(s): Lucie Guilbaud, Kévin Roger, Andree Schmidt, Cerina Chhuon, Stephan… http://dlvr.it/THCC1G #MassSpecRSS

Source: Journal of Proteomics
Author(s): Lucie Guilbaud, Kévin Roger, Andree Schmidt, Cerina Chhuon, Stephan… http://dlvr.it/THCC1G #MassSpecRSS
"...with the peptide sequences measured confirming the presence of South American camelids, most likely llama or alpaca."

"...with the peptide sequences measured confirming the presence of South American camelids, most likely llama or alpaca."


Human cell #proteomics subcellular landscape and its dynamic remodeling using #organelle IP.
Paper here www.cell.com/cell/fulltex...
Explore the date organelles.czbiohub.org
Congratulations to the team

Human cell #proteomics subcellular landscape and its dynamic remodeling using #organelle IP.
Paper here www.cell.com/cell/fulltex...
Explore the date organelles.czbiohub.org
Congratulations to the team

—Proteins associated with social isolation or loneliness and 5 diseases (left below) w/ increased risk
—Genes associated with social activities (right below) w/ protection


—Proteins associated with social isolation or loneliness and 5 diseases (left below) w/ increased risk
—Genes associated with social activities (right below) w/ protection
Glover, Schreiner, Dewson & Tait reviewed the intersection of #mitochondria and cell death. #pyroptosis #ferroptosis #necroptosis #apoptosis
👉https://rdcu.be/d46gt
www.nature.com/articles/s41...

Glover, Schreiner, Dewson & Tait reviewed the intersection of #mitochondria and cell death. #pyroptosis #ferroptosis #necroptosis #apoptosis
👉https://rdcu.be/d46gt
www.nature.com/articles/s41...



