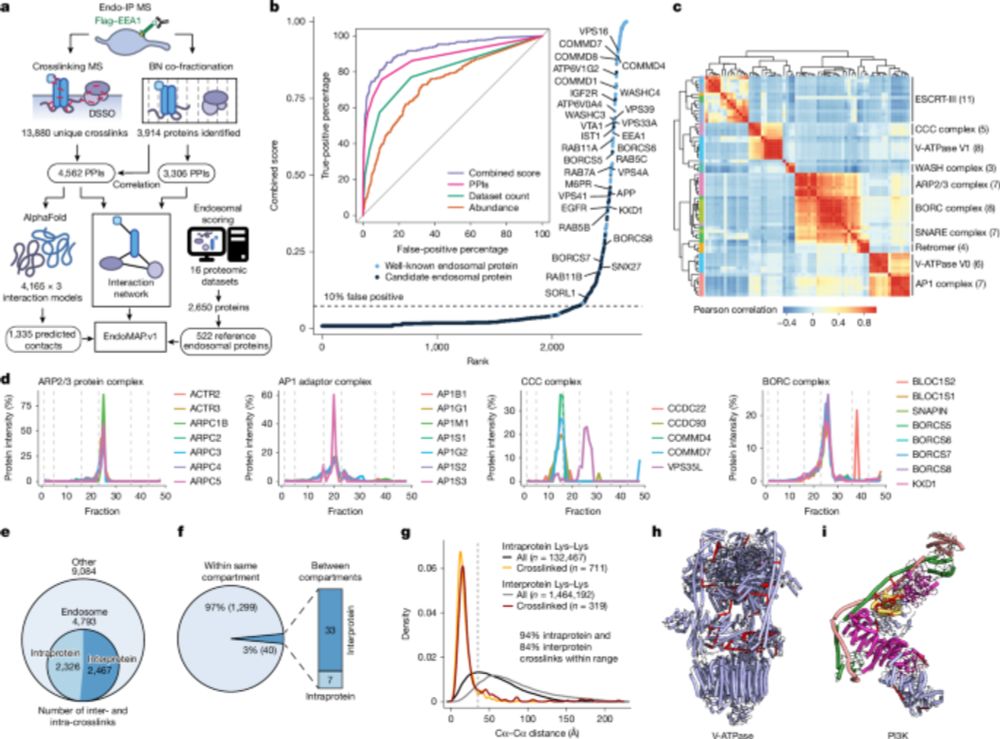


chemrxiv.org/engage/chemr...
chemrxiv.org/engage/chemr...
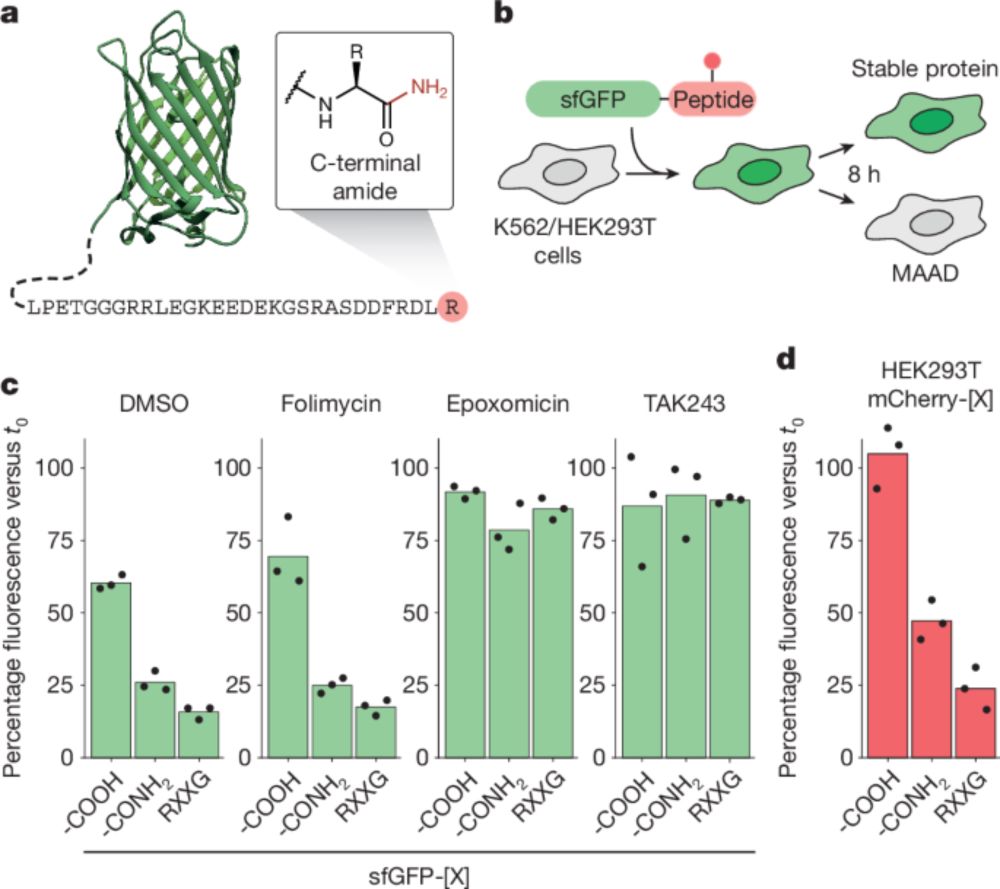
Excited to share our review article in @molcell.bsky.social, diving deep into everything #ubiquitin
Read here 👉: kwnsfk27.r.eu-west-1.awstrack.me/L0/https:%2F...
@cellpress.bsky.social
@wehi-research.bsky.social
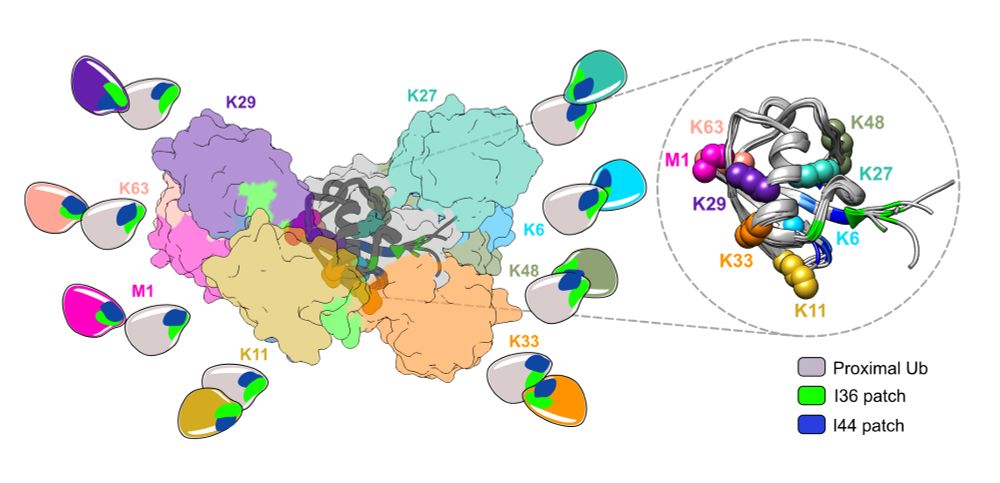
Excited to share our review article in @molcell.bsky.social, diving deep into everything #ubiquitin
Read here 👉: kwnsfk27.r.eu-west-1.awstrack.me/L0/https:%2F...
@cellpress.bsky.social
@wehi-research.bsky.social
www.biorxiv.org/content/10.1...
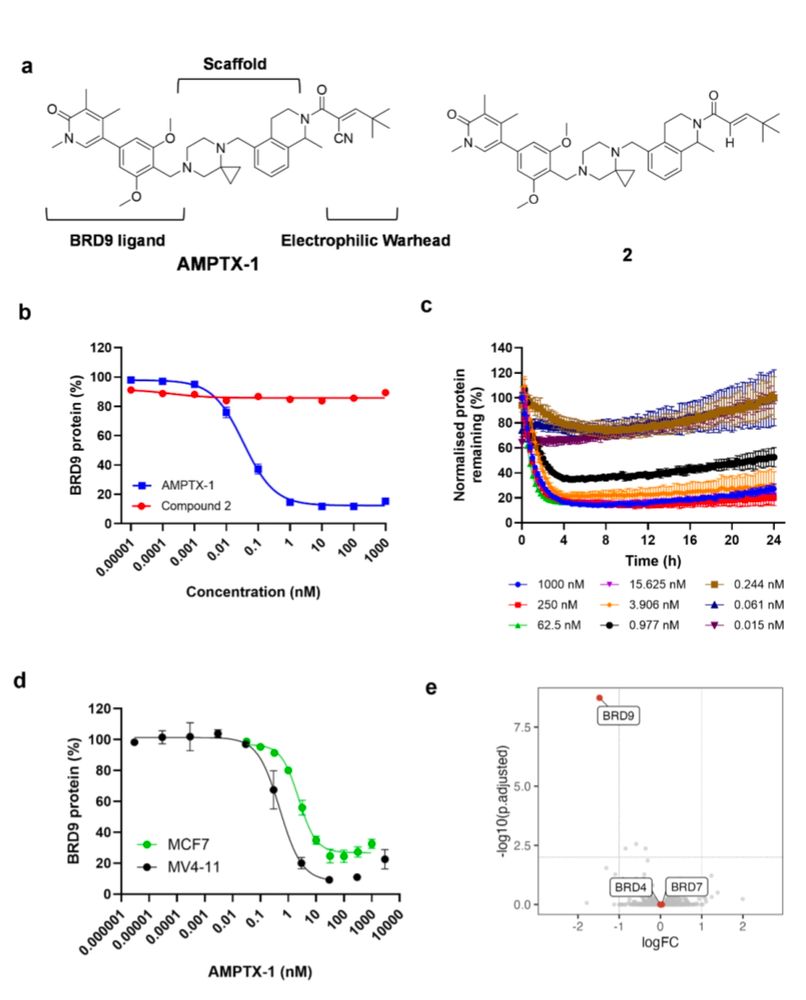
www.biorxiv.org/content/10.1...
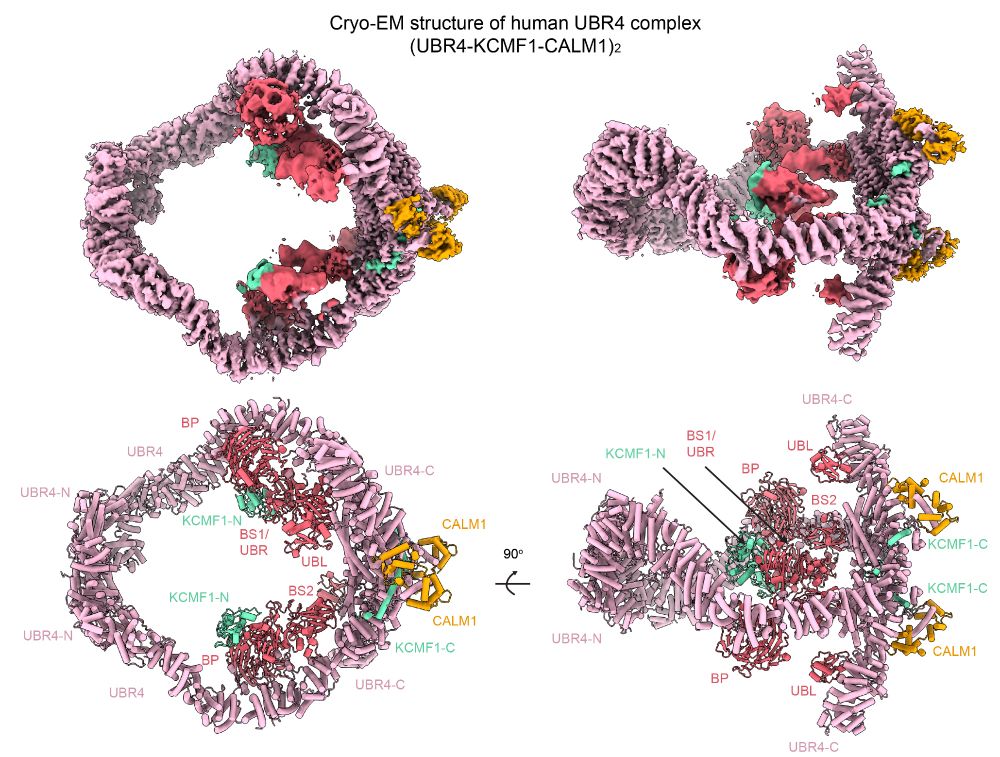
pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....

www.cell.com/molecular-ce...
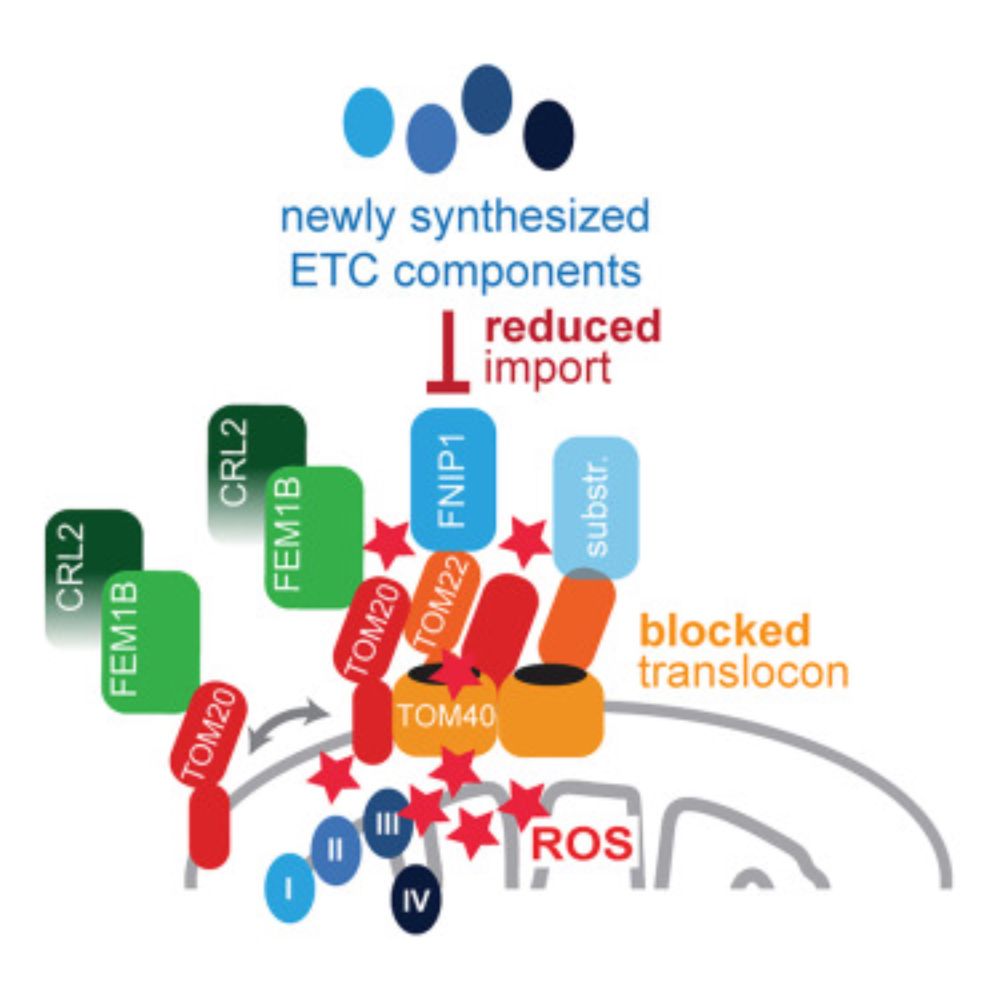
www.cell.com/molecular-ce...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


Have a look at the Ubiquitin and Ubls list Lisa :-)
Have a look at the Ubiquitin and Ubls list Lisa :-)


