Daniel Tamarit
@danieltamarit.bsky.social
Evolutionary biologist studying archaeal and bacterial genomes | Assistant Prof @BinfUtrecht.bsky.social 🇳🇱
Reposted by Daniel Tamarit
📢 Interested in #chromosomes 🧬 & #archaea thriving at the edge of life? PhD project in York,UK on how archaea pass on their genome to daughter cells.Friendly supervisors: me, @steve-quinn-lab.bsky.social & @georgerheath.bsky.social
www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!
www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!


November 9, 2025 at 5:20 PM
📢 Interested in #chromosomes 🧬 & #archaea thriving at the edge of life? PhD project in York,UK on how archaea pass on their genome to daughter cells.Friendly supervisors: me, @steve-quinn-lab.bsky.social & @georgerheath.bsky.social
www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!
www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!
Reposted by Daniel Tamarit
🧬 Registration is open for the Phylogenomics Workshop in Český Krumlov!
Fantastic training opportunity for anyone working with genomic data and evolutionary questions.
Highly recommended - amazing course, great community, and a fantastic setting!
Fantastic training opportunity for anyone working with genomic data and evolutionary questions.
Highly recommended - amazing course, great community, and a fantastic setting!

October 30, 2025 at 4:41 PM
🧬 Registration is open for the Phylogenomics Workshop in Český Krumlov!
Fantastic training opportunity for anyone working with genomic data and evolutionary questions.
Highly recommended - amazing course, great community, and a fantastic setting!
Fantastic training opportunity for anyone working with genomic data and evolutionary questions.
Highly recommended - amazing course, great community, and a fantastic setting!
Reposted by Daniel Tamarit
Delighted to be awarded NWO VIDI grant! We will study how microbes shape root cells to enhance plant stress resilience - so exciting!!!!
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
149 Vidi applications granted to talented researchers | NWO
NWO has granted a maximum of 850.000 euros to 149 scientists from the domain Science (ENW), Applied and Engineering Sciences (AES) and Social Sciences and Humanities (SSH) domains, as well as Health, Research and Development (ZonMw). With the help of the Vidi grant, the talented scientists can start their own line of research and further develop their talent.
www.nwo.nl
October 24, 2025 at 4:26 PM
Delighted to be awarded NWO VIDI grant! We will study how microbes shape root cells to enhance plant stress resilience - so exciting!!!!
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
Reposted by Daniel Tamarit
Incredibly exited to see @dorotakawa.bsky.social be awarded the "Dutch starting grant" (VIDI)! 🎉
And what a cohort, so many plant biologists awarded! Including @andrekuhn.bsky.social @charlieunderwood.bsky.social @kinpanchung.bsky.social - what a boost to the community and exciting science planned!
And what a cohort, so many plant biologists awarded! Including @andrekuhn.bsky.social @charlieunderwood.bsky.social @kinpanchung.bsky.social - what a boost to the community and exciting science planned!
Delighted to be awarded NWO VIDI grant! We will study how microbes shape root cells to enhance plant stress resilience - so exciting!!!!
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
www.nwo.nl/en/news/149-...
Fantastic to see several plant projects granted🌽🌱
149 Vidi applications granted to talented researchers | NWO
NWO has granted a maximum of 850.000 euros to 149 scientists from the domain Science (ENW), Applied and Engineering Sciences (AES) and Social Sciences and Humanities (SSH) domains, as well as Health, Research and Development (ZonMw). With the help of the Vidi grant, the talented scientists can start their own line of research and further develop their talent.
www.nwo.nl
October 28, 2025 at 10:22 AM
Incredibly exited to see @dorotakawa.bsky.social be awarded the "Dutch starting grant" (VIDI)! 🎉
And what a cohort, so many plant biologists awarded! Including @andrekuhn.bsky.social @charlieunderwood.bsky.social @kinpanchung.bsky.social - what a boost to the community and exciting science planned!
And what a cohort, so many plant biologists awarded! Including @andrekuhn.bsky.social @charlieunderwood.bsky.social @kinpanchung.bsky.social - what a boost to the community and exciting science planned!
Reposted by Daniel Tamarit
The mystery deepens: how did all complex life on Earth originate? In this intriguing VPRO Tegenlicht episode (in Dutch), PhD student Max Raas from the @kopslab.bsky.social talks about his collaboration with Berend Snel. They study the evolution of cell division machinery in single-celled organisms.
Hoe is al het complexe leven op aarde eigenlijk ontstaan?

Wat DNA uit slootwater ons vertelt over jouw onbekende voorouders | VPRO Tegenlicht
YouTube video by VPRO Tegenlicht
youtu.be
October 28, 2025 at 8:32 AM
The mystery deepens: how did all complex life on Earth originate? In this intriguing VPRO Tegenlicht episode (in Dutch), PhD student Max Raas from the @kopslab.bsky.social talks about his collaboration with Berend Snel. They study the evolution of cell division machinery in single-celled organisms.
Reposted by Daniel Tamarit
Our review is out in Nature Reviews Genetics! rdcu.be/d5AY2
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
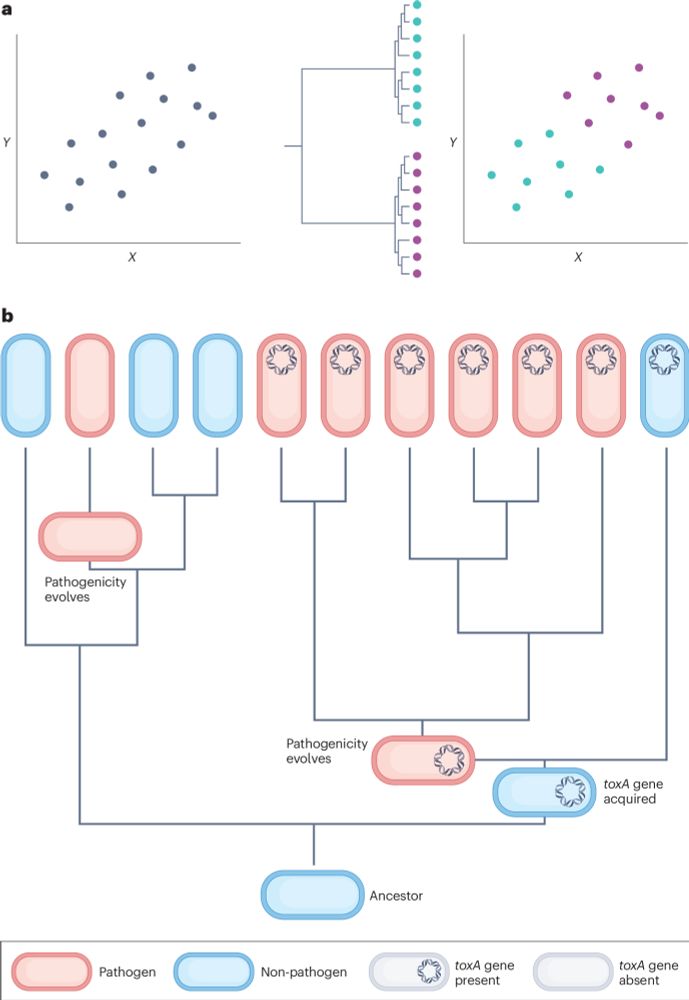
A phylogenetic approach to comparative genomics
Nature Reviews Genetics - Controlling for phylogeny is essential in comparative genomics studies, because species, genomes and genes are not independent data points within statistical tests. The...
rdcu.be
January 8, 2025 at 1:19 PM
Our review is out in Nature Reviews Genetics! rdcu.be/d5AY2
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
Reposted by Daniel Tamarit
S. Braichenko, @ruiborges23.bsky.social and C. Kosiol publish a new GBE Perspective on the potential applications of deep learning approaches in phylogenetics.
🔗 doi.org/10.1093/gbe/evaf177
#genome #evolution #phylogenetics
🔗 doi.org/10.1093/gbe/evaf177
#genome #evolution #phylogenetics

Phylogenetic Methods Meet Deep Learning
Abstract. Deep learning (DL) has been widely used in various scientific fields, but its integration into phylogenetics has been slower, primarily due to th
doi.org
October 27, 2025 at 11:07 AM
S. Braichenko, @ruiborges23.bsky.social and C. Kosiol publish a new GBE Perspective on the potential applications of deep learning approaches in phylogenetics.
🔗 doi.org/10.1093/gbe/evaf177
#genome #evolution #phylogenetics
🔗 doi.org/10.1093/gbe/evaf177
#genome #evolution #phylogenetics
Reposted by Daniel Tamarit
📢 We have an open position for a postdoc to join my lab. It's a great position @animecol-uu.bsky.social, fully salaried for 2.5 years with all benefits.
www.uu.se/en/about-uu/...
The project is about transmission patterns of bacteria and antimicrobial resistance (AMR) in aquatic insects. 🧬🦠 (1/3)
www.uu.se/en/about-uu/...
The project is about transmission patterns of bacteria and antimicrobial resistance (AMR) in aquatic insects. 🧬🦠 (1/3)

Postdoctoral researcher in molecular ecology - Uppsala University
Postdoctoral researcher in molecular ecology, Department of Ecology and Genetics, Uppsala University
www.uu.se
October 27, 2025 at 7:43 AM
📢 We have an open position for a postdoc to join my lab. It's a great position @animecol-uu.bsky.social, fully salaried for 2.5 years with all benefits.
www.uu.se/en/about-uu/...
The project is about transmission patterns of bacteria and antimicrobial resistance (AMR) in aquatic insects. 🧬🦠 (1/3)
www.uu.se/en/about-uu/...
The project is about transmission patterns of bacteria and antimicrobial resistance (AMR) in aquatic insects. 🧬🦠 (1/3)
Reposted by Daniel Tamarit
Glad to share our paper out today @NatureEcoEvo: “Serial innovations by Asgard archaea shaped the DNA replication machinery of the early eukaryotic ancestor”. www.nature.com/articles/s41... #microsky #archaeasky

Serial innovations by Asgard archaea shaped the DNA replication machinery of the early eukaryotic ancestor - Nature Ecology & Evolution
Phylogenetic and biochemical analyses show a diversity of components of the DNA replication machinery in different Asgard archaea that contributed to the eukaryotic DNA replication machinery.
www.nature.com
October 21, 2025 at 3:06 PM
Glad to share our paper out today @NatureEcoEvo: “Serial innovations by Asgard archaea shaped the DNA replication machinery of the early eukaryotic ancestor”. www.nature.com/articles/s41... #microsky #archaeasky
This is a beautiful study. Congrats Jose and all authors!
One of the most exciting works of my career, years in the making. We used high-throughput precision genome editing to test the fitness effects of thousands of natural variants. Our findings challenge the long-held assumption that common variants are inconsequential.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Massively parallel interrogation of the fitness of natural variants in ancient signaling pathways reveals pervasive local adaptation
The nature of standing genetic variation remains a central debate in population genetics, with differing perspectives on whether common variants are almost always neutral as suggested by neutral and n...
www.biorxiv.org
October 24, 2025 at 10:49 AM
This is a beautiful study. Congrats Jose and all authors!
Exultant to have been awarded a Vidi grant by the Dutch Research Council (NWO)! Can't wait to get started!
Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...
Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...

21 Vidi grants awarded to Utrecht-based researchers
21 researchers from Utrecht University, University Medical Center Utrecht, and Princess Máxima Center have each been awarded a Vidi grant worth up to €850,000.
www.uu.nl
October 23, 2025 at 12:49 PM
Exultant to have been awarded a Vidi grant by the Dutch Research Council (NWO)! Can't wait to get started!
Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...
Stay tuned for two upcoming (PhD student & postdoc) positions to study archaeal genome evolution.
Also huge congrats to my colleague @dorotakawa.bsky.social!
www.uu.nl/en/news/21-v...
Reposted by Daniel Tamarit
Scientists need more time to think and so do I. So do you.
Emails and messaging are the OG brainrot, and scrolling makes it so much worse.
I've spent a lot of coaching time with clients on creating more distance from their phone, literally and figuratively. Much benefit.
Emails and messaging are the OG brainrot, and scrolling makes it so much worse.
I've spent a lot of coaching time with clients on creating more distance from their phone, literally and figuratively. Much benefit.

October 23, 2025 at 7:00 AM
Scientists need more time to think and so do I. So do you.
Emails and messaging are the OG brainrot, and scrolling makes it so much worse.
I've spent a lot of coaching time with clients on creating more distance from their phone, literally and figuratively. Much benefit.
Emails and messaging are the OG brainrot, and scrolling makes it so much worse.
I've spent a lot of coaching time with clients on creating more distance from their phone, literally and figuratively. Much benefit.
Reposted by Daniel Tamarit
EdgeHOG reconstructs ancestral gene order at Tree-of-Life scale in linear time. 2,845 genomes → 1,133 ancestors. Vontigs for LECA (~1.8 Ga). Also dates gene adjacencies & rearrangements. Sexual chroms stand out! Led by Charles Bernard (research.pasteur.fr/fr/member/ch...) doi.org/10.1038/s415...

October 23, 2025 at 6:19 AM
EdgeHOG reconstructs ancestral gene order at Tree-of-Life scale in linear time. 2,845 genomes → 1,133 ancestors. Vontigs for LECA (~1.8 Ga). Also dates gene adjacencies & rearrangements. Sexual chroms stand out! Led by Charles Bernard (research.pasteur.fr/fr/member/ch...) doi.org/10.1038/s415...
Reposted by Daniel Tamarit
Testing the "least-diverged ortholog” conjecture on >1M structs + expression across 16 animals & 20 plants, the LDO tends to retain ancestral function while the other copy specialises. Work led by @irenejulca.bsky.social. Paper: doi.org/10.1101/gr.2... Free preprint: www.biorxiv.org/content/10.1...

October 23, 2025 at 6:17 AM
Testing the "least-diverged ortholog” conjecture on >1M structs + expression across 16 animals & 20 plants, the LDO tends to retain ancestral function while the other copy specialises. Work led by @irenejulca.bsky.social. Paper: doi.org/10.1101/gr.2... Free preprint: www.biorxiv.org/content/10.1...
Reposted by Daniel Tamarit
Structure-informed phylogenetic inference with FoldTree outperforms sequence-only trees on divergent proteins, and untangles RRNPPA quorum-sensing receptors across Gram-positive bacteria & phages. Work led by David Moi (www.linkedin.com/in/david-moi) Paper: doi.org/10.1038/s415...

October 23, 2025 at 6:13 AM
Structure-informed phylogenetic inference with FoldTree outperforms sequence-only trees on divergent proteins, and untangles RRNPPA quorum-sensing receptors across Gram-positive bacteria & phages. Work led by David Moi (www.linkedin.com/in/david-moi) Paper: doi.org/10.1038/s415...
Reposted by Daniel Tamarit
Our @narjournal.bsky.social manuscript is out! It explores the growth of the GTDB (gtdb.ecogenomic.org) since its inception, as well as updates to the website, methodology, policies, and major taxonomic and nomenclatural changes over the past three years.
academic.oup.com/nar/advance-...
academic.oup.com/nar/advance-...

GTDB release 10: a complete and systematic taxonomy for 715 230 bacterial and 17 245 archaeal genomes
Abstract. The Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) provides a phylogenetically consistent and rank normalized genome-based taxonomy
academic.oup.com
October 22, 2025 at 2:20 PM
Our @narjournal.bsky.social manuscript is out! It explores the growth of the GTDB (gtdb.ecogenomic.org) since its inception, as well as updates to the website, methodology, policies, and major taxonomic and nomenclatural changes over the past three years.
academic.oup.com/nar/advance-...
academic.oup.com/nar/advance-...
Reposted by Daniel Tamarit
One of the most exciting works of my career, years in the making. We used high-throughput precision genome editing to test the fitness effects of thousands of natural variants. Our findings challenge the long-held assumption that common variants are inconsequential.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Massively parallel interrogation of the fitness of natural variants in ancient signaling pathways reveals pervasive local adaptation
The nature of standing genetic variation remains a central debate in population genetics, with differing perspectives on whether common variants are almost always neutral as suggested by neutral and n...
www.biorxiv.org
October 22, 2025 at 5:46 PM
One of the most exciting works of my career, years in the making. We used high-throughput precision genome editing to test the fitness effects of thousands of natural variants. Our findings challenge the long-held assumption that common variants are inconsequential.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Daniel Tamarit
Featuring Paulien Hogeweg (2nd from the right), founder and lifelong colleague at @binfutrecht.bsky.social @utrechtuniversity.bsky.social
Very well deserved, Paulien!
Very well deserved, Paulien!
October 22, 2025 at 2:06 PM
Featuring Paulien Hogeweg (2nd from the right), founder and lifelong colleague at @binfutrecht.bsky.social @utrechtuniversity.bsky.social
Very well deserved, Paulien!
Very well deserved, Paulien!
Reposted by Daniel Tamarit
A fun little side project I've been working on with @stepadenisov.bsky.social , Mato Lagator, and Andreas Wagner: "Strong promoters are mutationally robust". Briefly...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Strong promoters are mutationally robust
Mutational robustness is the persistence of a phenotype upon mutation. It facilitates molecular evolution and has been characterized in a variety of biological systems, but studies of prokaryotic prom...
www.biorxiv.org
October 21, 2025 at 8:02 AM
A fun little side project I've been working on with @stepadenisov.bsky.social , Mato Lagator, and Andreas Wagner: "Strong promoters are mutationally robust". Briefly...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Daniel Tamarit
Lunch today halfway around the world in Hangzhou and met a new asst professor who was very thankful for this resource. Please help out folks like them and other early career scientists with examples of job apps. Submit yours!
github.com/RILAB/statem...
github.com/RILAB/statem...

GitHub - RILAB/statements: Successful Job Applications and Grants
Successful Job Applications and Grants. Contribute to RILAB/statements development by creating an account on GitHub.
github.com
October 21, 2025 at 6:55 AM
Lunch today halfway around the world in Hangzhou and met a new asst professor who was very thankful for this resource. Please help out folks like them and other early career scientists with examples of job apps. Submit yours!
github.com/RILAB/statem...
github.com/RILAB/statem...
Reposted by Daniel Tamarit
JOB ALERT! We are looking for a LAB MANAGER (7-year position!) to help us build the MOLECULAR BIODIVERSITY lab in our @terra-cluster.org at @unituebingen.bsky.social. Are you a NGS wet lab expert, and you like the idea of supporting research on natural biodiversity, then apply!
Please re-post!
Please re-post!

October 17, 2025 at 11:51 AM
JOB ALERT! We are looking for a LAB MANAGER (7-year position!) to help us build the MOLECULAR BIODIVERSITY lab in our @terra-cluster.org at @unituebingen.bsky.social. Are you a NGS wet lab expert, and you like the idea of supporting research on natural biodiversity, then apply!
Please re-post!
Please re-post!
Reposted by Daniel Tamarit
Hot off the press! Our latest work on the evolution of facultative symbiosis in stony corals, focusing on a remarkable Mediterranean species: Oculina patagonica.
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...


October 15, 2025 at 8:29 PM
Hot off the press! Our latest work on the evolution of facultative symbiosis in stony corals, focusing on a remarkable Mediterranean species: Oculina patagonica.
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
Reposted by Daniel Tamarit
Hello everyone, take a look at my cool bugs from the Microbial Dark Matter phylum Saccharibacteria! These ultrasmall bacteria (in green) track down Actinobacteria hosts (not in green) and grow on the host cell envelope.
For the first time, scientists have performed targeted mutations on an episymbiosis-determining pathway. In a new study on Saccharibacteria, AFI researchers used advanced techniques to observe pili that drive motility and host attachment.
@batbilegbor.bsky.social
forsyth.org/saccharibact...
@batbilegbor.bsky.social
forsyth.org/saccharibact...
October 15, 2025 at 5:01 PM
Hello everyone, take a look at my cool bugs from the Microbial Dark Matter phylum Saccharibacteria! These ultrasmall bacteria (in green) track down Actinobacteria hosts (not in green) and grow on the host cell envelope.
Reposted by Daniel Tamarit
We are happy to share our latest work in @nature.com . We study the genomic and cellular basis of facultative symbiosis in Oculina patagonica - a Mediterranean coral remarkable for its ability to survive long periods without algal symbionts. Led by Shani Levy and @xgrau.bsky.social
rdcu.be/eLbaZ
rdcu.be/eLbaZ

October 15, 2025 at 7:58 PM
We are happy to share our latest work in @nature.com . We study the genomic and cellular basis of facultative symbiosis in Oculina patagonica - a Mediterranean coral remarkable for its ability to survive long periods without algal symbionts. Led by Shani Levy and @xgrau.bsky.social
rdcu.be/eLbaZ
rdcu.be/eLbaZ
Reposted by Daniel Tamarit
Excited to share our new work on nanopore metabarcoding — showing how it can be used for protists and combined with short reads!
Led by my PhD student Małgorzata Chwalińska, with our team at @ibe-warszawa.bsky.social and @fabnot.bsky.social & Sarah Romac at @sbroscoff.bsky.social
#ProtistsOnSky
Led by my PhD student Małgorzata Chwalińska, with our team at @ibe-warszawa.bsky.social and @fabnot.bsky.social & Sarah Romac at @sbroscoff.bsky.social
#ProtistsOnSky
"Want to start using Nanopore technology to research protistan diversity? Check out our paper introducing a pipeline for creating OTUs from Nanopore metabarcodes — bridging the gap between short- and long-read metabarcoding." - Anna Karnkowska
doi.org/10.3897/mbmg...
doi.org/10.3897/mbmg...

October 14, 2025 at 2:53 PM
Excited to share our new work on nanopore metabarcoding — showing how it can be used for protists and combined with short reads!
Led by my PhD student Małgorzata Chwalińska, with our team at @ibe-warszawa.bsky.social and @fabnot.bsky.social & Sarah Romac at @sbroscoff.bsky.social
#ProtistsOnSky
Led by my PhD student Małgorzata Chwalińska, with our team at @ibe-warszawa.bsky.social and @fabnot.bsky.social & Sarah Romac at @sbroscoff.bsky.social
#ProtistsOnSky


