
https://biochem.emory.edu/bou-nader-lab/
#BYI #beckman #science #grants #academia #chemistry #lifesciences




genesdev.cshlp.org/cgi/content/...
genesdev.cshlp.org/cgi/content/...
academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.science.org/doi/10.1126/...
@unmc.bsky.social @uwbiochem.bsky.social

www.science.org/doi/10.1126/...
@unmc.bsky.social @uwbiochem.bsky.social
We describe a new tetrameric RAD51 paralog complex – XRCC3-RAD51C-RAD51D-XRCC2 – which caps the end of RAD51 filaments.
Link: www.science.org/doi/epdf/10....
Thread ⬇️ (1/8)
We describe a new tetrameric RAD51 paralog complex – XRCC3-RAD51C-RAD51D-XRCC2 – which caps the end of RAD51 filaments.
Link: www.science.org/doi/epdf/10....
Thread ⬇️ (1/8)
@wyssinstitute.bsky.social
journals.asm.org/doi/10.1128/...

@wyssinstitute.bsky.social
journals.asm.org/doi/10.1128/...
Abbey Vito’s first publication discusses a tug-of-war between BER enzymes and chromatin for access to damaged DNA, a battle central to genome stability.
Built on Tyler's foundational work! tinyurl.com/ywkt2bt5

Abbey Vito’s first publication discusses a tug-of-war between BER enzymes and chromatin for access to damaged DNA, a battle central to genome stability.
Built on Tyler's foundational work! tinyurl.com/ywkt2bt5


The Human RNome Project has been launched: a global effort to map all human RNAs and their chemical modifications. Proud to support it and contribute to the article in Genome Biology doi.org/10.1186/s130...
#RNA #bioinformatics #RNAstructure #modomics

The Human RNome Project has been launched: a global effort to map all human RNAs and their chemical modifications. Proud to support it and contribute to the article in Genome Biology doi.org/10.1186/s130...
#RNA #bioinformatics #RNAstructure #modomics

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
pubs.acs.org/doi/10.1021/acscentsci.5c01345

pubs.acs.org/doi/10.1021/acscentsci.5c01345
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
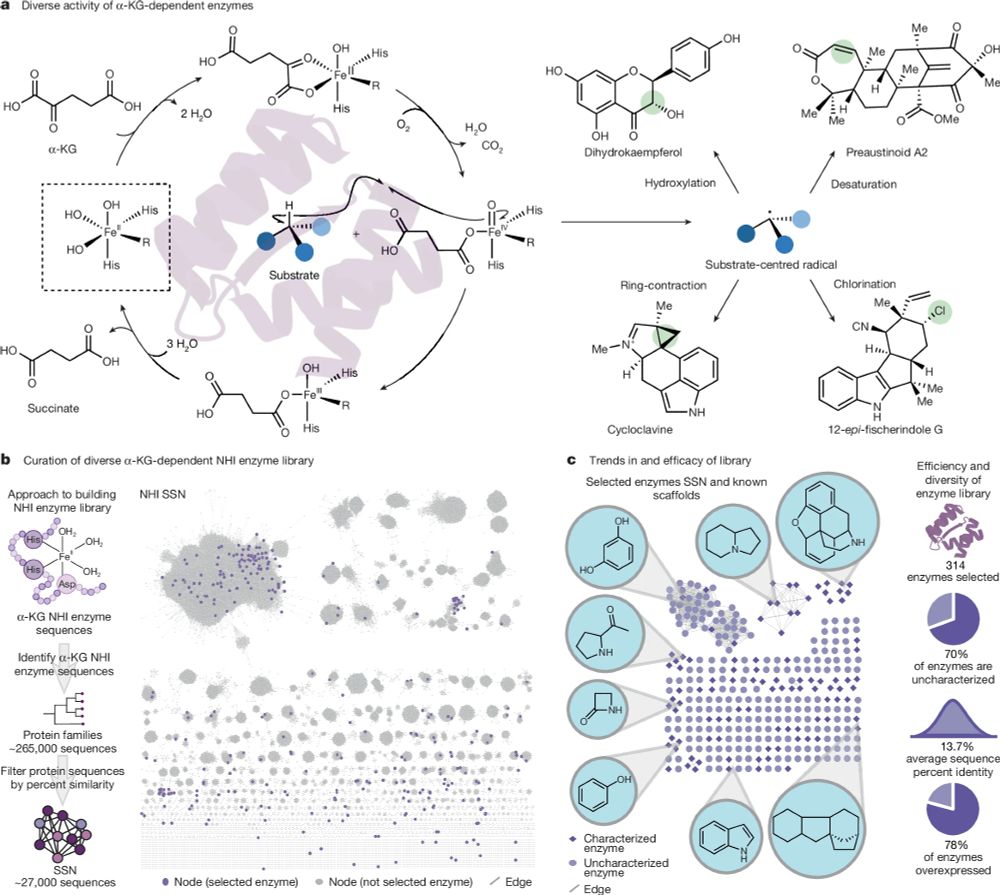

@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...

