Ben Good
@benjaminhgood.bsky.social
Assistant Professor of Applied Physics @Stanford. Theoretical biophysics. Evolutionary dynamics. Microbial evolution. https://bgoodlab.github.io/
Pinned
Ben Good
@benjaminhgood.bsky.social
· Aug 16

Prehistoric Global Migration of Vanishing Gut Microbes With Humans
The gut microbiome is crucial for health and greatly affected by lifestyle. Many microbes common in non-industrialized populations are disappearing or extinct in industrialized populations. Understand...
www.biorxiv.org
Excited to share a new preprint w/ the Sonnenberg lab, led by Matt Carter, @zzzhiru.bsky.social & @mattolm.bsky.social. We analyzed the microbiomes of two non-industrialized populations from opposite sides of the globe to try to reconstruct the recent evolutionary history of our gut microbiota.
Reposted by Ben Good
🚨 Microbiologists! We are recruiting Assistant / Associate Professors in 3 collaborative areas of our U. Pittsburgh School of Medicine.
1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...
1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...

Faculty Professor Associate - Full-Time | Vaughn Cooper
We are recruiting Faculty microbiologists in three (3) different, complementary, and collaborative areas at the University of Pittsburgh associated with the School of Medicine.
1) Fundamental researc...
www.linkedin.com
September 23, 2025 at 10:31 PM
🚨 Microbiologists! We are recruiting Assistant / Associate Professors in 3 collaborative areas of our U. Pittsburgh School of Medicine.
1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...
1) MMG (my dept): fundamental research in med micro
2) Peds ID / I4Kids institute
3) Center for Vaccine Research
🔗 to all 3 w/info: www.linkedin.com/posts/vaughn...
Reposted by Ben Good
Now hiring a computational postdoc (evolutionary genomics, molecular evolution) in my lab at Emory University.
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...
Careers | Emory University | Atlanta GA
faculty-emory.icims.com
September 18, 2025 at 3:38 PM
Now hiring a computational postdoc (evolutionary genomics, molecular evolution) in my lab at Emory University.
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...
If you’re interested in population genetics, fitness landscapes, and viral evolution — get in touch.
faculty-emory.icims.com/jobs/151181/...
Reposted by Ben Good
The constant barrage of terrible news on bluesky has made me feel weird about promoting papers, but people in the lab have been doing so much amazing work over the past few months that I want to share a few brief teasers/links:
September 10, 2025 at 4:46 PM
The constant barrage of terrible news on bluesky has made me feel weird about promoting papers, but people in the lab have been doing so much amazing work over the past few months that I want to share a few brief teasers/links:
Excited to share a new preprint w/ the Sonnenberg lab, led by Matt Carter, @zzzhiru.bsky.social & @mattolm.bsky.social. We analyzed the microbiomes of two non-industrialized populations from opposite sides of the globe to try to reconstruct the recent evolutionary history of our gut microbiota.

Prehistoric Global Migration of Vanishing Gut Microbes With Humans
The gut microbiome is crucial for health and greatly affected by lifestyle. Many microbes common in non-industrialized populations are disappearing or extinct in industrialized populations. Understand...
www.biorxiv.org
August 16, 2025 at 6:25 PM
Excited to share a new preprint w/ the Sonnenberg lab, led by Matt Carter, @zzzhiru.bsky.social & @mattolm.bsky.social. We analyzed the microbiomes of two non-industrialized populations from opposite sides of the globe to try to reconstruct the recent evolutionary history of our gut microbiota.
Reposted by Ben Good
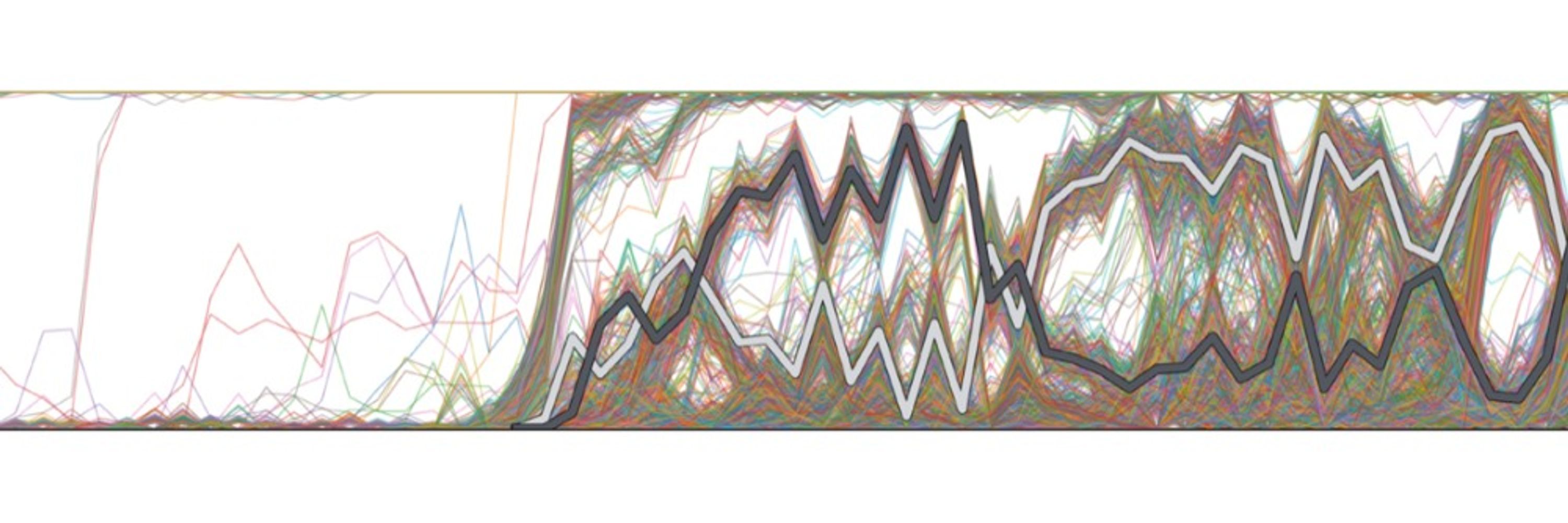
Incredibly proud of this work where we developed a method for understanding the information contained in millions of genomes. Another example of NIH funded research.
📣Online NOW!
📄Estimation of demography and mutation rates from one million haploid genomes
🧑🤝🧑 @jgschraiber.bsky.social @jeffspence.github.io @docedge.bsky.social
📄Estimation of demography and mutation rates from one million haploid genomes
🧑🤝🧑 @jgschraiber.bsky.social @jeffspence.github.io @docedge.bsky.social
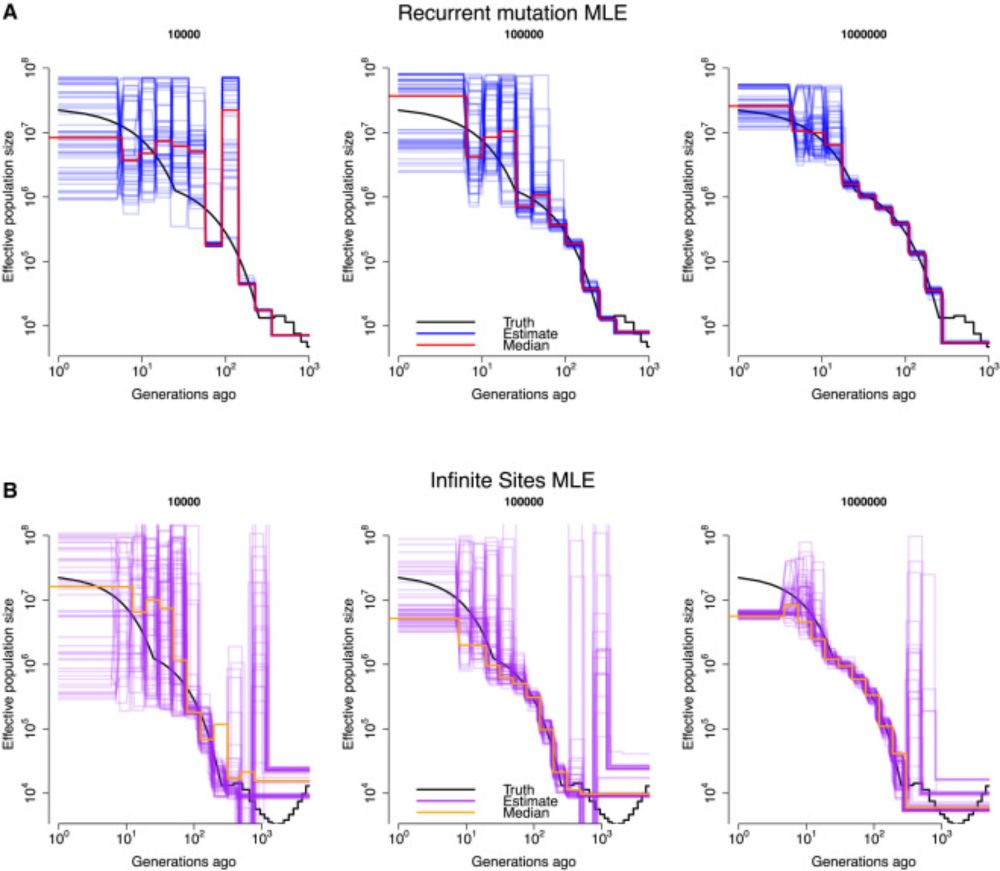
Estimation of demography and mutation rates from one million haploid genomes
Samples of millions of genomes provide substantial information about recent demography
and mutation, but standard population-genetic methods make assumptions not met in
these data. We introduce DR EVI...
www.cell.com
August 13, 2025 at 2:47 PM
Incredibly proud of this work where we developed a method for understanding the information contained in millions of genomes. Another example of NIH funded research.
Reposted by Ben Good
You can get an accurate estimate of total bacterial biomass from stool metagenomes by simply normalizing by host read count, without needing any additional measurements.
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Metagenomic estimation of absolute bacterial biomass in the mammalian gut through host-derived read normalization | mSystems
In this study, we asked whether normalization by host reads alone was sufficient to
estimate absolute bacterial biomass directly from stool metagenomic data, without
the need for synthetic spike-ins, ...
journals.asm.org
July 31, 2025 at 3:33 PM
You can get an accurate estimate of total bacterial biomass from stool metagenomes by simply normalizing by host read count, without needing any additional measurements.
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Reposted by Ben Good
I'm thrilled that my lab at NYU is now supported by an NIH MIRA grant! I'm looking to hire 1-2 senior lab members (outstanding postdoc candidates or experienced staff scientists) with expertise in computational or statistical methods in human genetics or genomics. Please share!
July 25, 2025 at 8:29 PM
I'm thrilled that my lab at NYU is now supported by an NIH MIRA grant! I'm looking to hire 1-2 senior lab members (outstanding postdoc candidates or experienced staff scientists) with expertise in computational or statistical methods in human genetics or genomics. Please share!
Reposted by Ben Good
The Xue lab at UC Irvine is looking for a staff scientist to support our work investigating how microbes interact and evolve in the gut microbiome! Open to a wide range of previous experience levels, see ad for more.
recruit.ap.uci.edu/JPF09601
recruit.ap.uci.edu/JPF09601

Junior, Assistant, or Associate Specialist – Xue Lab
University of California, Irvine is hiring. Apply now!
recruit.ap.uci.edu
July 17, 2025 at 8:32 PM
The Xue lab at UC Irvine is looking for a staff scientist to support our work investigating how microbes interact and evolve in the gut microbiome! Open to a wide range of previous experience levels, see ad for more.
recruit.ap.uci.edu/JPF09601
recruit.ap.uci.edu/JPF09601
Reposted by Ben Good
Finding easy regions for short-read variant calling from pangenome data arxiv.org/abs/2507.03718 🧬🖥️🧪 github.com/lh3/panmask

July 11, 2025 at 6:30 PM
Finding easy regions for short-read variant calling from pangenome data arxiv.org/abs/2507.03718 🧬🖥️🧪 github.com/lh3/panmask
Reposted by Ben Good
Staff scientist position (computational):
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
July 7, 2025 at 3:27 PM
Staff scientist position (computational):
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
Reposted by Ben Good
Check out Laura's thread on our latest preprint demonstrating best methods for low biomass skin sampling.
Excited to share my new preprint: we show that for lab control and skin samples with low microbial biomass, metagenomics significantly outperforms 16S while DNA extraction method has minimal impact. 🧵 1/4
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Microbiome diversity of low biomass skin sites is captured by metagenomics but not 16S amplicon sequencing
Established workflows for microbiome analysis work well for high microbial biomass samples, like stool, but often fail to accurately define microbial communities when applied to low microbial biomass ...
www.biorxiv.org
June 25, 2025 at 12:36 PM
Check out Laura's thread on our latest preprint demonstrating best methods for low biomass skin sampling.
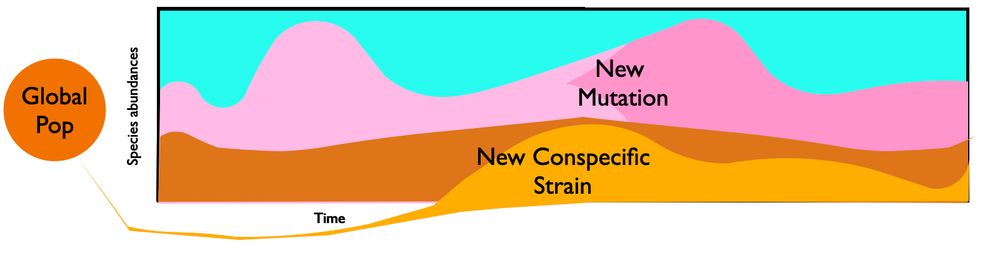
Next up is Sophie Walton, who will be talking about some new in vitro experiments to quantify the dynamics of selection among conspecific strains in larger microbial communities (Monday 9:30am, Evolutionary Ecology I).
Super excited for #Evol2025! I will be sharing some cool work on selection in the gut microbial communities at 9:30 am on Monday in the Evolutionary Ecology I session (Parthenon 2)!! Looking forward to chatting with folks as well!
June 21, 2025 at 4:14 AM
Next up is Sophie Walton, who will be talking about some new in vitro experiments to quantify the dynamics of selection among conspecific strains in larger microbial communities (Monday 9:30am, Evolutionary Ecology I).
Two members of our group will be presenting at #evolution2025 this year. First up is Anastasia Lyulina, who will be presenting some new theory on the site frequency spectrum of selected mutations in non-equilibrium populations (Saturday at 4:15; Population Genetics Theory IV).
Looking forward to #evolution2025! I will be talking about how time-varying demography and selection shape the site frequency spectrum — Saturday at 4:15 pm, Population Genetics Theory IV. Come say hi if you are around!
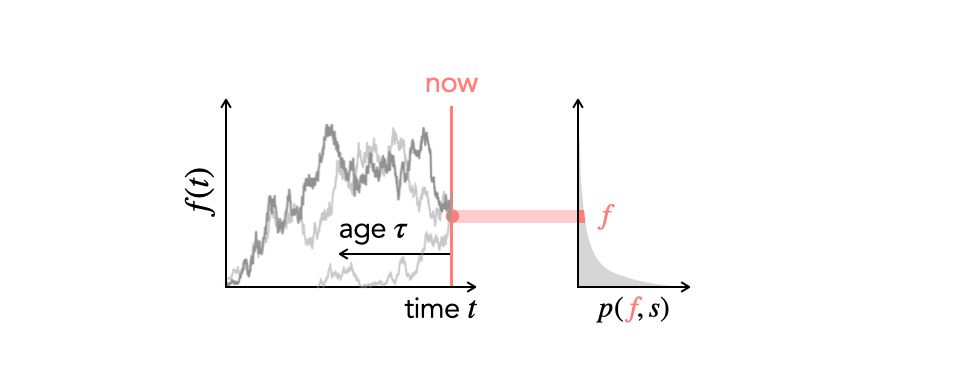
June 21, 2025 at 4:10 AM
Two members of our group will be presenting at #evolution2025 this year. First up is Anastasia Lyulina, who will be presenting some new theory on the site frequency spectrum of selected mutations in non-equilibrium populations (Saturday at 4:15; Population Genetics Theory IV).
Thanks for these questions @skryazhi.bsky.social! I might add that we’ve been playing around a bit with FGM-like models to see when you might expect behavior like this, and it turns out to be a lot more complex than (at least I) expected.
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
June 20, 2025 at 4:17 PM
Thanks for these questions @skryazhi.bsky.social! I might add that we’ve been playing around a bit with FGM-like models to see when you might expect behavior like this, and it turns out to be a lot more complex than (at least I) expected.
Reposted by Ben Good
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
June 20, 2025 at 1:34 PM
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
Reposted by Ben Good
There are some implicit assumptions that must be true for this pleiotropic expansion model to work. First, just to clarify, the fact that a box is colorful does not mean it has a positive effect on fitness, it just means it is relevant. So a mutant's affect on a trait can be good in E1, bad in E2
June 20, 2025 at 1:34 PM
There are some implicit assumptions that must be true for this pleiotropic expansion model to work. First, just to clarify, the fact that a box is colorful does not mean it has a positive effect on fitness, it just means it is relevant. So a mutant's affect on a trait can be good in E1, bad in E2
Reposted by Ben Good
Thanks for taking a look! We actually have an updated v2 manuscript that I think clarifies our two competing hypotheses in figure 1:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
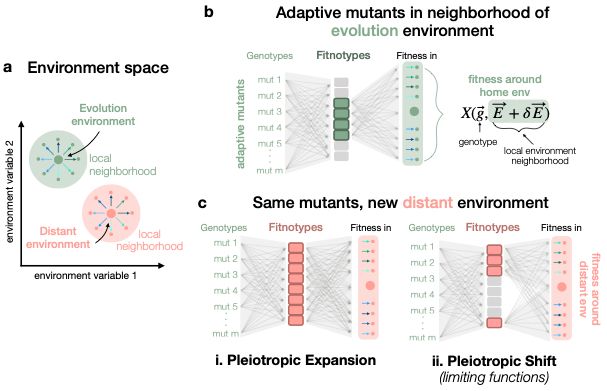
June 19, 2025 at 2:38 PM
Thanks for taking a look! We actually have an updated v2 manuscript that I think clarifies our two competing hypotheses in figure 1:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Ben Good
@oliviamghosh.bsky.social @grantkinsler.bsky.social @benjaminhgood.bsky.social @petrovadmitri.bsky.social: I am reading your “Low-dimensional genotype-fitness mapping ... ” preprint. Very interesting!
Can you help me understand the "pleiotropic expansion model" (Fig 1a)?
shorturl.at/qRYfJ
Can you help me understand the "pleiotropic expansion model" (Fig 1a)?
shorturl.at/qRYfJ
www.biorxiv.org
June 19, 2025 at 4:42 AM
@oliviamghosh.bsky.social @grantkinsler.bsky.social @benjaminhgood.bsky.social @petrovadmitri.bsky.social: I am reading your “Low-dimensional genotype-fitness mapping ... ” preprint. Very interesting!
Can you help me understand the "pleiotropic expansion model" (Fig 1a)?
shorturl.at/qRYfJ
Can you help me understand the "pleiotropic expansion model" (Fig 1a)?
shorturl.at/qRYfJ
Reposted by Ben Good
I have an opportunity to hire a staff scientist for my lab. Looking for someone with outstanding skillset in ML/statistics, genomics applications; interest in mentoring, strong publication record, PD experience required.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
June 1, 2025 at 3:33 PM
I have an opportunity to hire a staff scientist for my lab. Looking for someone with outstanding skillset in ML/statistics, genomics applications; interest in mentoring, strong publication record, PD experience required.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
Reposted by Ben Good
Super excited to release a huge evolution project on the works for many years:
Evolution experiments synchronized across climates to understand rapid adaptation
Preprint: doi.org/10.1101/2025...
All data available: www.grene-net.org/data
#MOILAB
@ucberkeleyofficial.bsky.social
@hhmi.org
🧵👇
Evolution experiments synchronized across climates to understand rapid adaptation
Preprint: doi.org/10.1101/2025...
All data available: www.grene-net.org/data
#MOILAB
@ucberkeleyofficial.bsky.social
@hhmi.org
🧵👇
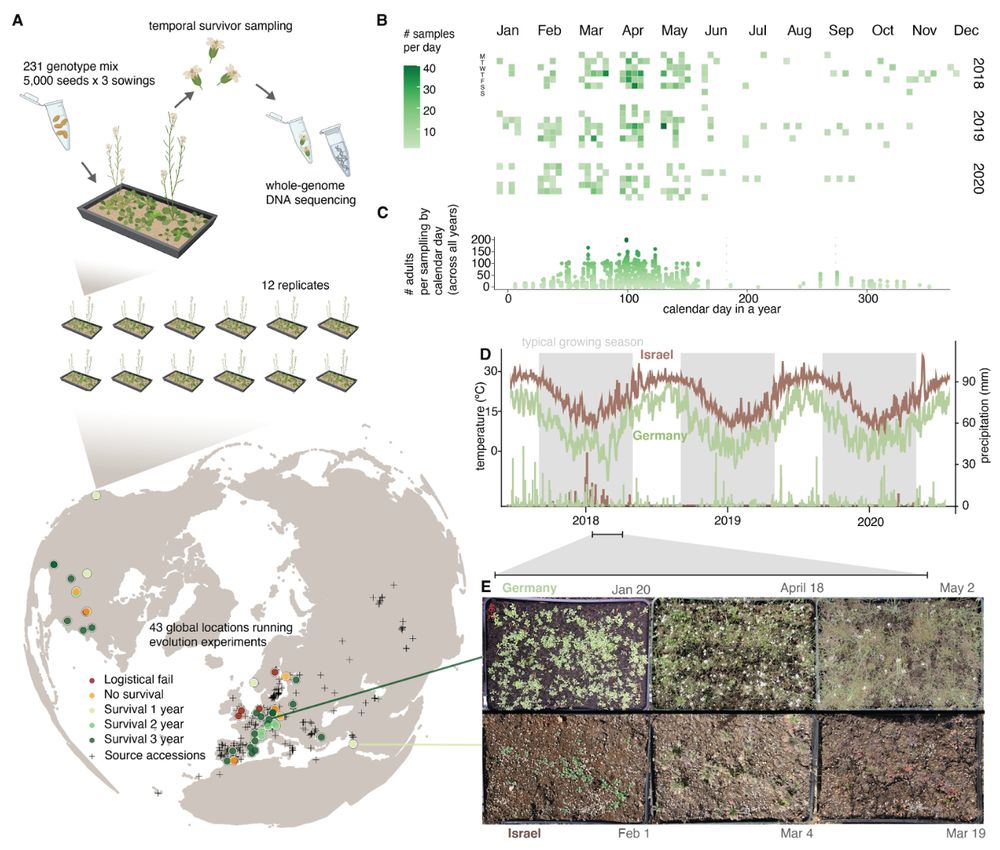
May 30, 2025 at 5:56 PM
Super excited to release a huge evolution project on the works for many years:
Evolution experiments synchronized across climates to understand rapid adaptation
Preprint: doi.org/10.1101/2025...
All data available: www.grene-net.org/data
#MOILAB
@ucberkeleyofficial.bsky.social
@hhmi.org
🧵👇
Evolution experiments synchronized across climates to understand rapid adaptation
Preprint: doi.org/10.1101/2025...
All data available: www.grene-net.org/data
#MOILAB
@ucberkeleyofficial.bsky.social
@hhmi.org
🧵👇
Reposted by Ben Good
1/n 🧵 Excited to share our new paper! We developed a framework to reveal hidden simplicity in how organisms adapt to different environments, particularly focusing on antibiotic resistance evolution. #EvolutionaryBiology #MachineLearning
Learning the Shape of Evolutionary Landscapes: Geometric Deep Learning Reveals Hidden Structure in Phenotype-to-Fitness Maps https://www.biorxiv.org/content/10.1101/2025.05.07.652616v1
May 15, 2025 at 2:33 PM
1/n 🧵 Excited to share our new paper! We developed a framework to reveal hidden simplicity in how organisms adapt to different environments, particularly focusing on antibiotic resistance evolution. #EvolutionaryBiology #MachineLearning
Reposted by Ben Good
The most profound insights are often also the most simple:
growth-rate, by setting the dilution rate of intra-cellular molecules, controls the sensitivity of gene regulatory circuits. In retrospect it seems crazy that this effect seems to have been overlooked so far.
www.science.org/doi/10.1126/...
growth-rate, by setting the dilution rate of intra-cellular molecules, controls the sensitivity of gene regulatory circuits. In retrospect it seems crazy that this effect seems to have been overlooked so far.
www.science.org/doi/10.1126/...
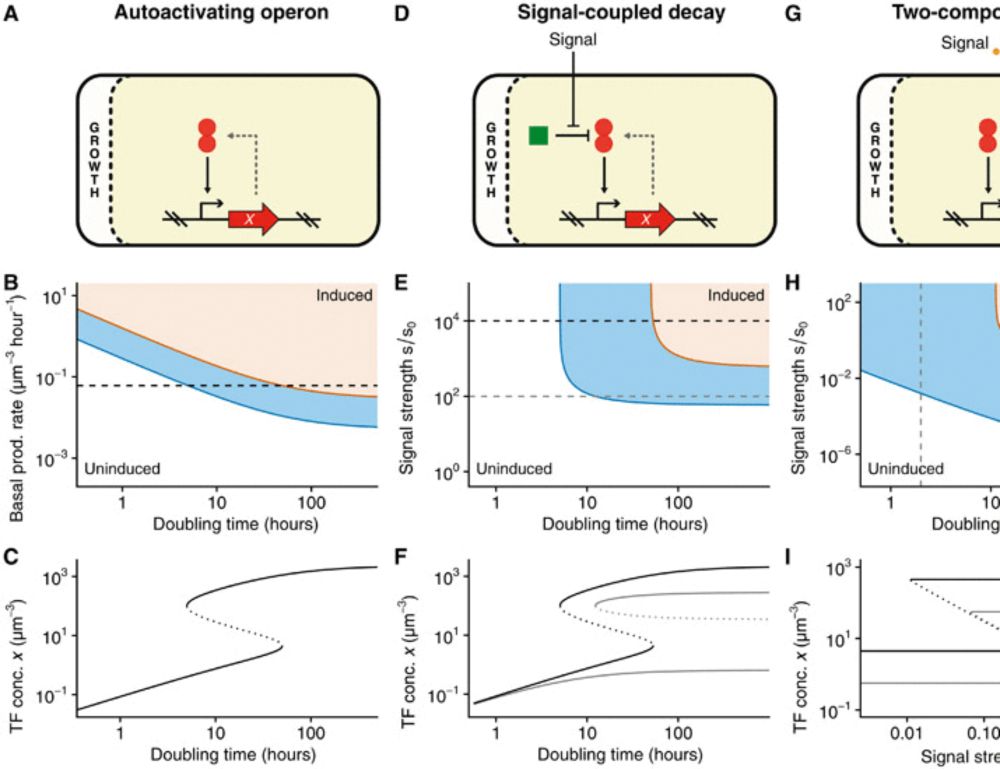
Growth rate controls the sensitivity of gene regulatory circuits
Through the simple effect of dilution rate, regulatory circuits systematically change their sensitivity with growth rate.
www.science.org
April 29, 2025 at 1:28 PM
The most profound insights are often also the most simple:
growth-rate, by setting the dilution rate of intra-cellular molecules, controls the sensitivity of gene regulatory circuits. In retrospect it seems crazy that this effect seems to have been overlooked so far.
www.science.org/doi/10.1126/...
growth-rate, by setting the dilution rate of intra-cellular molecules, controls the sensitivity of gene regulatory circuits. In retrospect it seems crazy that this effect seems to have been overlooked so far.
www.science.org/doi/10.1126/...
Reposted by Ben Good
I’m thrilled to share my first ever publication, now published in PNAS! www.pnas.org/doi/10.1073/...
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
www.pnas.org
March 11, 2025 at 1:22 PM
I’m thrilled to share my first ever publication, now published in PNAS! www.pnas.org/doi/10.1073/...
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
Reposted by Ben Good
Legitmately thrilled to share our latest work, in which @fernpizza.bsky.social solved an experimental challenge in plasmid biology as old as the field: measuring how plasmids compete and evolve within individual cells!
February 21, 2025 at 8:42 PM
Legitmately thrilled to share our latest work, in which @fernpizza.bsky.social solved an experimental challenge in plasmid biology as old as the field: measuring how plasmids compete and evolve within individual cells!


