Ben Good
@benjaminhgood.bsky.social
Assistant Professor of Applied Physics @Stanford. Theoretical biophysics. Evolutionary dynamics. Microbial evolution. https://bgoodlab.github.io/
Interestingly, of the subset of species shared w/ industrialized pop'ns, some show evidence of recent transmission, while others suggest a more ancient association. Among these, the genetic isolation between Tsimane & Hadza strains was generally larger than between comparable industrialized pop'ns

August 17, 2025 at 9:59 PM
Interestingly, of the subset of species shared w/ industrialized pop'ns, some show evidence of recent transmission, while others suggest a more ancient association. Among these, the genetic isolation between Tsimane & Hadza strains was generally larger than between comparable industrialized pop'ns
We were surprised to see right off the bat that - despite their long history of geographic separation - most of the microbial species in the Tsimane microbiome were also found in the Hadza (and most of these are either rare or absent in industrialized populations).

August 17, 2025 at 9:43 PM
We were surprised to see right off the bat that - despite their long history of geographic separation - most of the microbial species in the Tsimane microbiome were also found in the Hadza (and most of these are either rare or absent in industrialized populations).
Excited to share some new work led by John McEnany. We generalize random matrix approaches from community assembly theory to predict how the fitness benefits & fates of new mutations should scale with the diversity & metabolic overlap of their surrounding community. We'd love to hear your comments!

December 18, 2023 at 8:42 PM
Excited to share some new work led by John McEnany. We generalize random matrix approaches from community assembly theory to predict how the fitness benefits & fates of new mutations should scale with the diversity & metabolic overlap of their surrounding community. We'd love to hear your comments!
Yeah I agree – it’s still a bit of a mystery for me too! Our paper suggests that the simplest spatial models can’t lower the Ne by *too* much in the human gut, but it’s possible that unmodeled aspects of spatial structure could contribute to this (e.g. diurnal oscillations)

August 24, 2023 at 8:19 PM
Yeah I agree – it’s still a bit of a mystery for me too! Our paper suggests that the simplest spatial models can’t lower the Ne by *too* much in the human gut, but it’s possible that unmodeled aspects of spatial structure could contribute to this (e.g. diurnal oscillations)
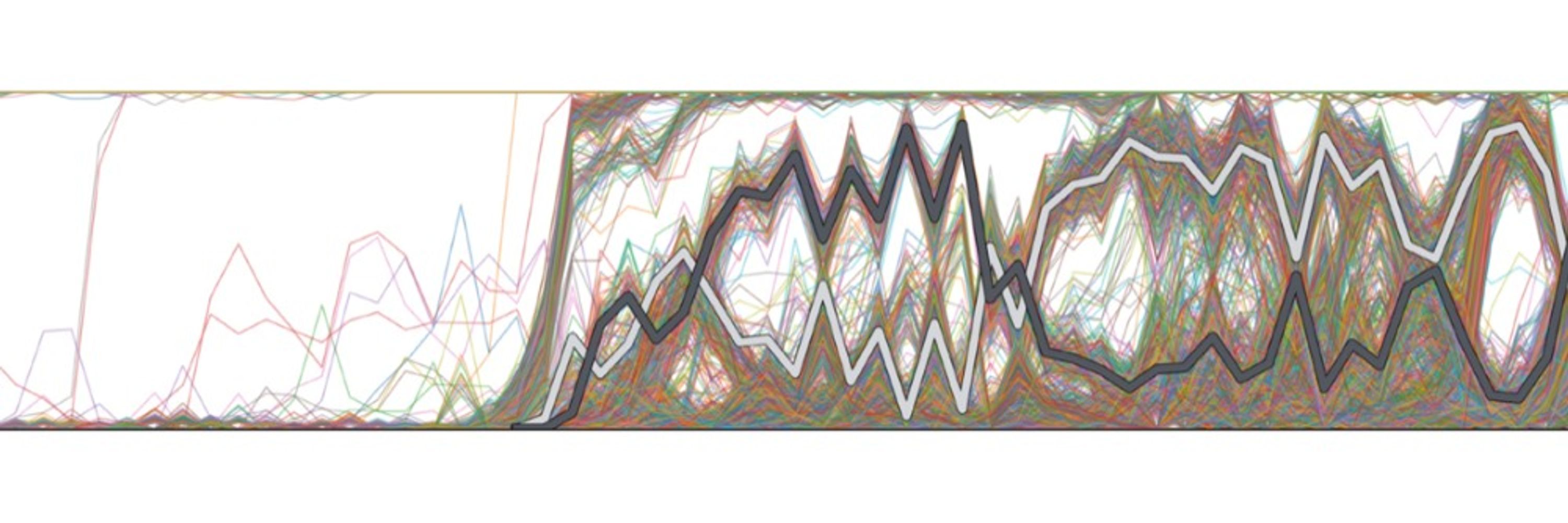
If it helps, we also looked into this question in Fig 3 of this paper: www.pnas.org/doi/10.1073/... (similar model to the paper above). The answer turns out to depend on selection strength - more strongly beneficial mut'ns can arise in a larger effective pop'n (not always = to # of dividing cells)

August 24, 2023 at 3:09 PM
If it helps, we also looked into this question in Fig 3 of this paper: www.pnas.org/doi/10.1073/... (similar model to the paper above). The answer turns out to depend on selection strength - more strongly beneficial mut'ns can arise in a larger effective pop'n (not always = to # of dividing cells)

