Ben Good
@benjaminhgood.bsky.social
Assistant Professor of Applied Physics @Stanford. Theoretical biophysics. Evolutionary dynamics. Microbial evolution. https://bgoodlab.github.io/
Drifting into speculation: I think these data support the picture that gut microbes can spread much faster than human genes. Strong geographic barriers can be still be very important (as in the Tsimane & Hadza case) but the magnitude of this effect
stands out compared to other contemporary pop'ns.
stands out compared to other contemporary pop'ns.

Limited codiversification of the gut microbiota with humans
A recent study by Suzuki & Fitzstevens et al ([1][1]) argued that dozens of species of gut bacteria have codiversified with modern human populations. Reanalysis of their data reveals that the correlat...
www.biorxiv.org
August 17, 2025 at 10:45 PM
Drifting into speculation: I think these data support the picture that gut microbes can spread much faster than human genes. Strong geographic barriers can be still be very important (as in the Tsimane & Hadza case) but the magnitude of this effect
stands out compared to other contemporary pop'ns.
stands out compared to other contemporary pop'ns.
Interestingly, of the subset of species shared w/ industrialized pop'ns, some show evidence of recent transmission, while others suggest a more ancient association. Among these, the genetic isolation between Tsimane & Hadza strains was generally larger than between comparable industrialized pop'ns

August 17, 2025 at 9:59 PM
Interestingly, of the subset of species shared w/ industrialized pop'ns, some show evidence of recent transmission, while others suggest a more ancient association. Among these, the genetic isolation between Tsimane & Hadza strains was generally larger than between comparable industrialized pop'ns
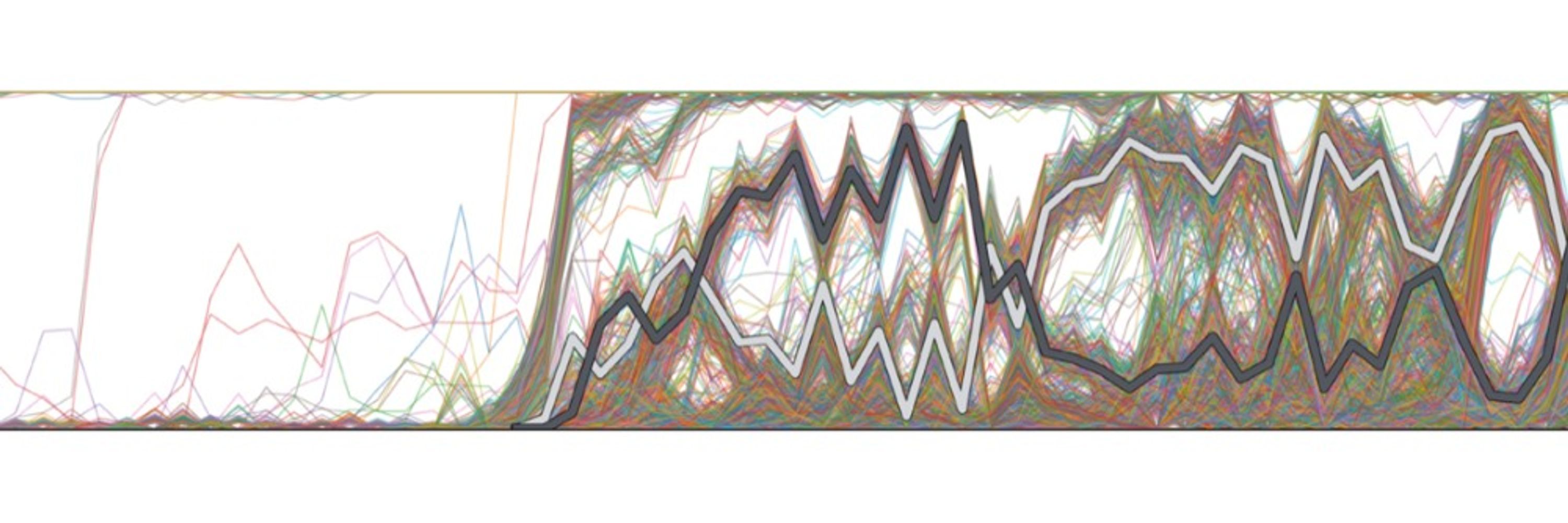
We developed several methods to detect & date the onset of genetic isolation between these microbial pop'ns, which leverage the fact that gut bacteria constantly exchange DNA w/ each other. In most species, we found patterns of divergence & gene flow consistent w/ prehistoric co-migration w/ humans.

Dynamics of bacterial recombination in the human gut microbiome
Recombination is a ubiquitous force in bacterial evolution, but its dynamics are poorly characterized in many natural populations. This study presents a metagenomic approach for quantifying the landsc...
journals.plos.org
August 17, 2025 at 9:43 PM
We developed several methods to detect & date the onset of genetic isolation between these microbial pop'ns, which leverage the fact that gut bacteria constantly exchange DNA w/ each other. In most species, we found patterns of divergence & gene flow consistent w/ prehistoric co-migration w/ humans.
We were surprised to see right off the bat that - despite their long history of geographic separation - most of the microbial species in the Tsimane microbiome were also found in the Hadza (and most of these are either rare or absent in industrialized populations).

August 17, 2025 at 9:43 PM
We were surprised to see right off the bat that - despite their long history of geographic separation - most of the microbial species in the Tsimane microbiome were also found in the Hadza (and most of these are either rare or absent in industrialized populations).
Benefiting from decades of work by the Tsimane Life History Project (tsimane.anth.ucsb.edu) and a previous set of samples collected by Melanie Martin, @danielsprockett.bsky.social & co, we used deep metagenomic sequencing to reconstruct ~12k microbial genomes from ~80 Tsimane adults.
The UNM-UCSB Tsimane Health and Life History Project
The UNM-Tsimane Health and Life History Project
tsimane.anth.ucsb.edu
August 17, 2025 at 9:43 PM
Benefiting from decades of work by the Tsimane Life History Project (tsimane.anth.ucsb.edu) and a previous set of samples collected by Melanie Martin, @danielsprockett.bsky.social & co, we used deep metagenomic sequencing to reconstruct ~12k microbial genomes from ~80 Tsimane adults.
Here we tried to get around this challenge by analyzing two geographically distant populations – the Tsimane of Bolivia and the Hadza of Tanzania – whose ancestors have been separated for more than 10,000 years prior to recent human globalization.
August 17, 2025 at 9:43 PM
Here we tried to get around this challenge by analyzing two geographically distant populations – the Tsimane of Bolivia and the Hadza of Tanzania – whose ancestors have been separated for more than 10,000 years prior to recent human globalization.
Why am I so excited about this work? There’s been a lot of interest in understanding how gut bacteria might have evolved (or co-evolved) w/ humans. But the plasticity of the microbiome can make it difficult to distinguish long-term associations from more recent acquisitions from the environment.
August 17, 2025 at 9:43 PM
Why am I so excited about this work? There’s been a lot of interest in understanding how gut bacteria might have evolved (or co-evolved) w/ humans. But the plasticity of the microbiome can make it difficult to distinguish long-term associations from more recent acquisitions from the environment.
I think this emphasizes the need for more concrete models of pleiotropy to help us know when these verbal models should even apply in principle (let alone in expts like @oliviamghosh.bsky.social's). In that vein, was very excited about the new work that @djhelam1.bsky.social talked about at NITMB
June 20, 2025 at 4:27 PM
I think this emphasizes the need for more concrete models of pleiotropy to help us know when these verbal models should even apply in principle (let alone in expts like @oliviamghosh.bsky.social's). In that vein, was very excited about the new work that @djhelam1.bsky.social talked about at NITMB
Reposted by Ben Good
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
June 20, 2025 at 1:34 PM
I think it would be super cool to explore some of these assumptions and models theoretically, but (spoiler alert) we don't actually find evidence for the pleiotropic expansion model in our data! So perhaps these assumptions are not borne out in reality (in our system at least).
Reposted by Ben Good
There are some implicit assumptions that must be true for this pleiotropic expansion model to work. First, just to clarify, the fact that a box is colorful does not mean it has a positive effect on fitness, it just means it is relevant. So a mutant's affect on a trait can be good in E1, bad in E2
June 20, 2025 at 1:34 PM
There are some implicit assumptions that must be true for this pleiotropic expansion model to work. First, just to clarify, the fact that a box is colorful does not mean it has a positive effect on fitness, it just means it is relevant. So a mutant's affect on a trait can be good in E1, bad in E2

