Proud to see strong #EORTC representation with six abstracts and a joint session with BTG featuring EORTC-led projects on clinical trial planning.
More info 👉 www.eortc.org/blog/2025/10...
#EANO2025 #NeuroOncology @qol.eortc.org

Proud to see strong #EORTC representation with six abstracts and a joint session with BTG featuring EORTC-led projects on clinical trial planning.
More info 👉 www.eortc.org/blog/2025/10...
#EANO2025 #NeuroOncology @qol.eortc.org
@fnr.lu, #precisionmedicine, #organoids, #drugscreen, #pharmacogenomics, #braintumors

@fnr.lu, #precisionmedicine, #organoids, #drugscreen, #pharmacogenomics, #braintumors


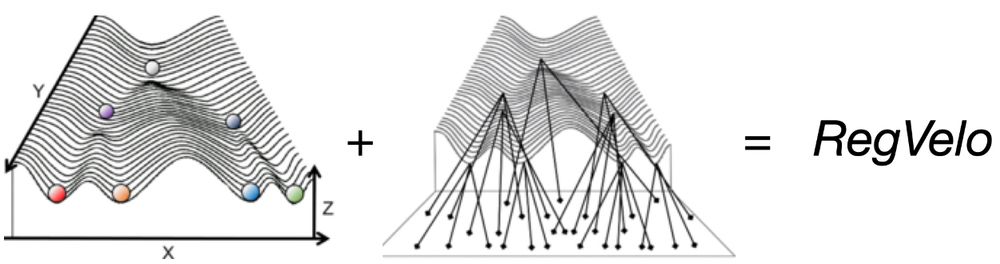
doi.org/10.1093/neuo...

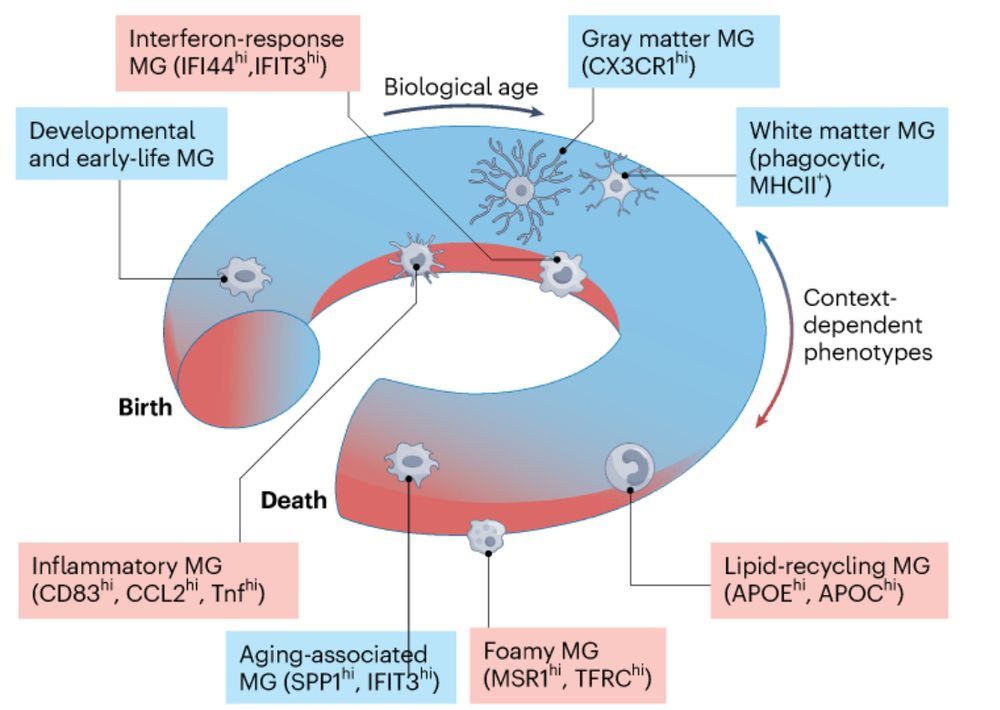
Great people, great science.
#glioblastoma #spatialtranscriptomics
#scRNA-seq




Great people, great science.
#glioblastoma #spatialtranscriptomics
#scRNA-seq
Excited about the open positions as part of our newly funded Emmy Noether Group are online now under
kocakavuklab.vercel.app/joinus
📢 Please RT (what's the bsky equivalent here?) and distribute !
👇A quick instruction on how to apply
Excited about the open positions as part of our newly funded Emmy Noether Group are online now under
kocakavuklab.vercel.app/joinus
📢 Please RT (what's the bsky equivalent here?) and distribute !
👇A quick instruction on how to apply
https://go.nature.com/434f4ns

https://go.nature.com/434f4ns

www.cell.com/cell-reports...

@10xGenomics
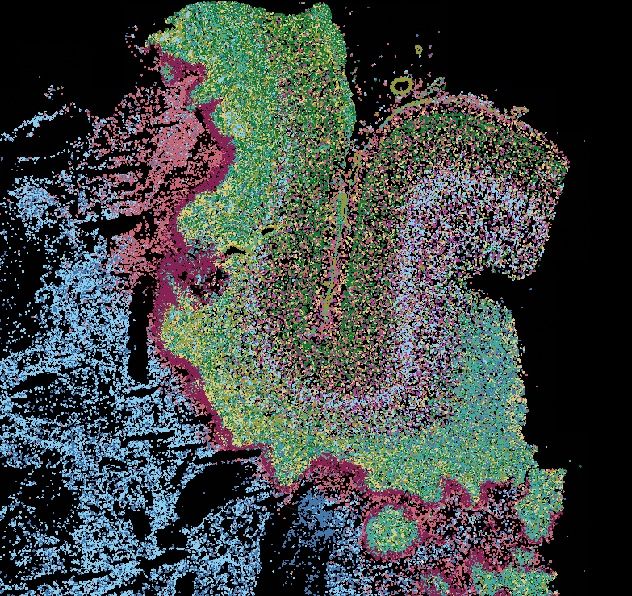
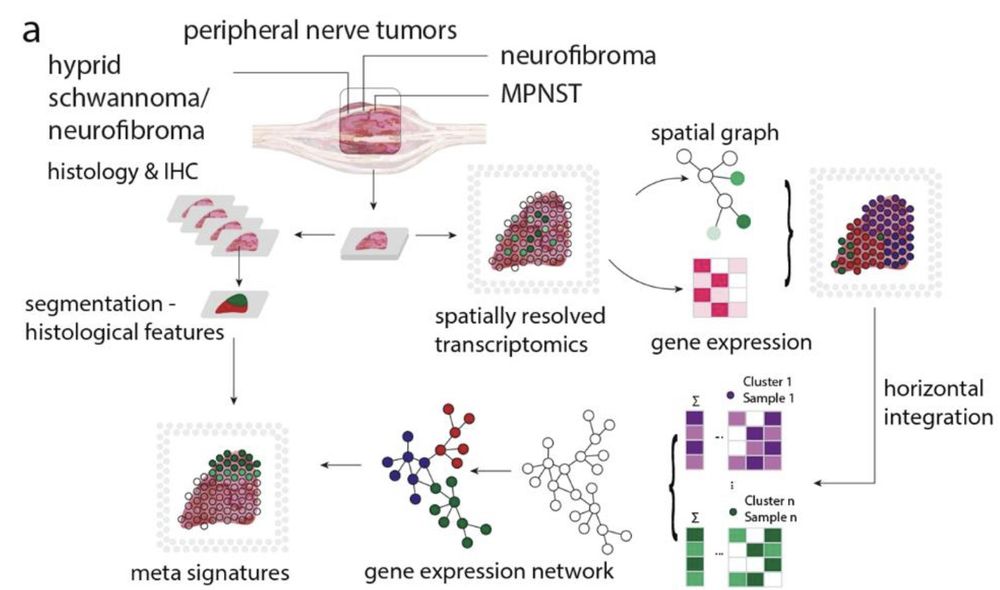
🚨Spoiler Alert: We identified APOD in neurofibroma regions as a novel biomarker in hybrid PNSTs
t.co/GmA9MLNKy1
@10xGenomics
🚨Spoiler Alert: We identified APOD in neurofibroma regions as a novel biomarker in hybrid PNSTs
t.co/GmA9MLNKy1
Excited by the breakthroughs in spatial transcriptomics and single-cell analysis? Follow @milolab.bsky.social account on Instagram: cutting-edge insights, updates, and engaging content!
www.instagram.com/the_milolab/...
Excited by the breakthroughs in spatial transcriptomics and single-cell analysis? Follow @milolab.bsky.social account on Instagram: cutting-edge insights, updates, and engaging content!
www.instagram.com/the_milolab/...
@sahm_lab
leveraging spatial transcriptomics
@10xGenomics
for neuropathology diagnosis #DeepLearning
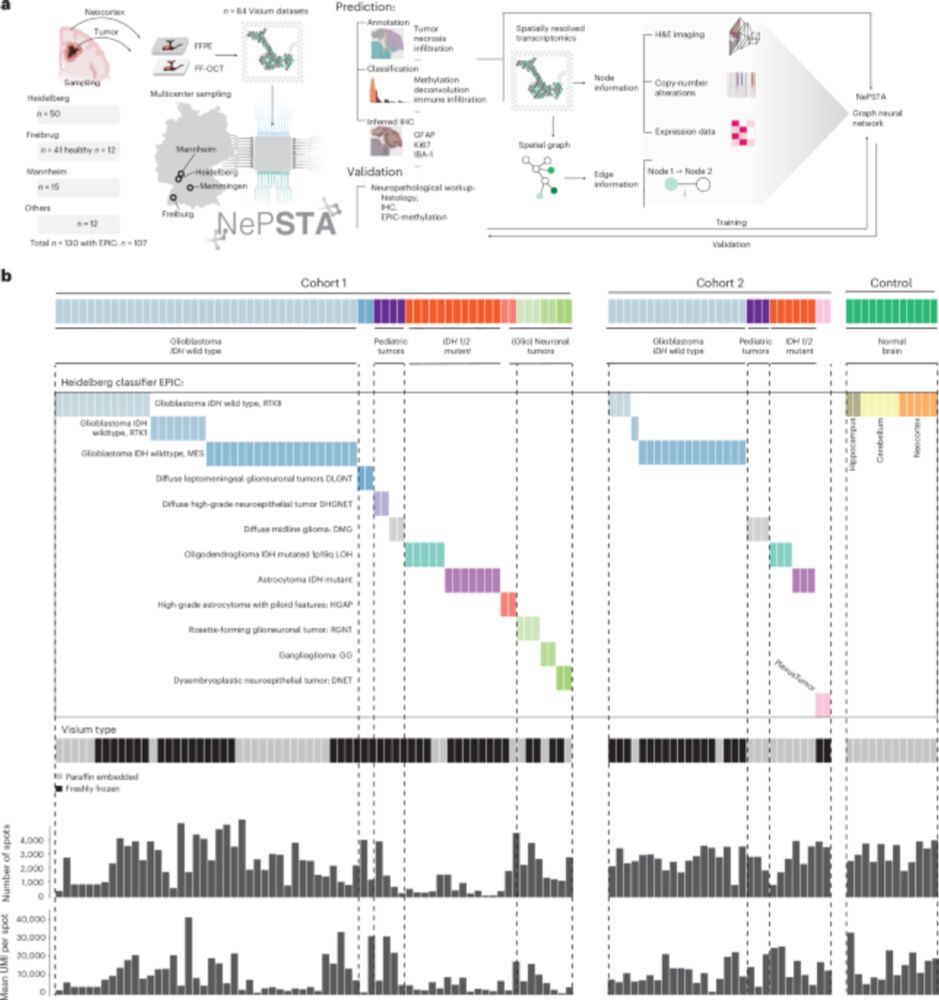
! 107 patients across 15 CNS tumors!
www.nature.com/articles/s43...
@sahm_lab
leveraging spatial transcriptomics
@10xGenomics
for neuropathology diagnosis #DeepLearning
! 107 patients across 15 CNS tumors!
www.nature.com/articles/s43...
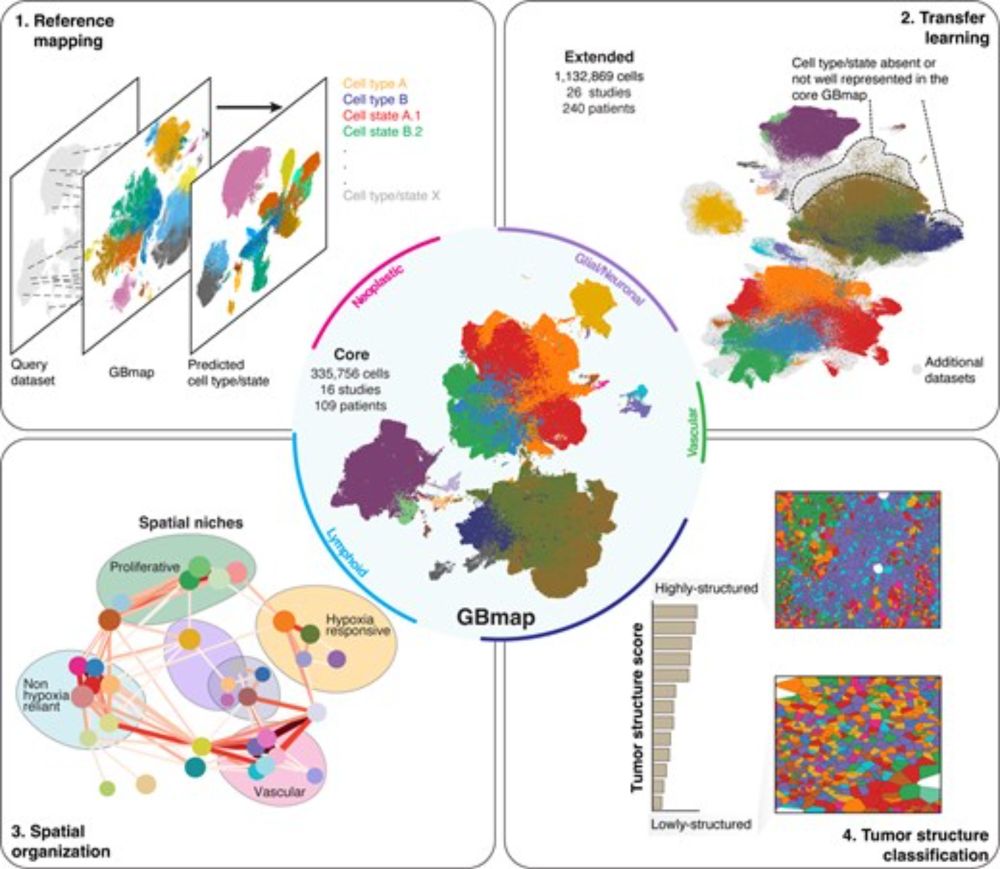
Using deep learning to enhance brain tumor diagnosis. From spatial transcriptomics data you can predict tissue histology, methylation subclass, genotype, immunohistochemistry etc.
Using deep learning to enhance brain tumor diagnosis. From spatial transcriptomics data you can predict tissue histology, methylation subclass, genotype, immunohistochemistry etc.
Spatially resolved transcriptomics and graph-based deep learning improve accuracy of routine CNS tumor diagnostics | @naturecancer.bsky.social
www.nature.com/articles/s43...
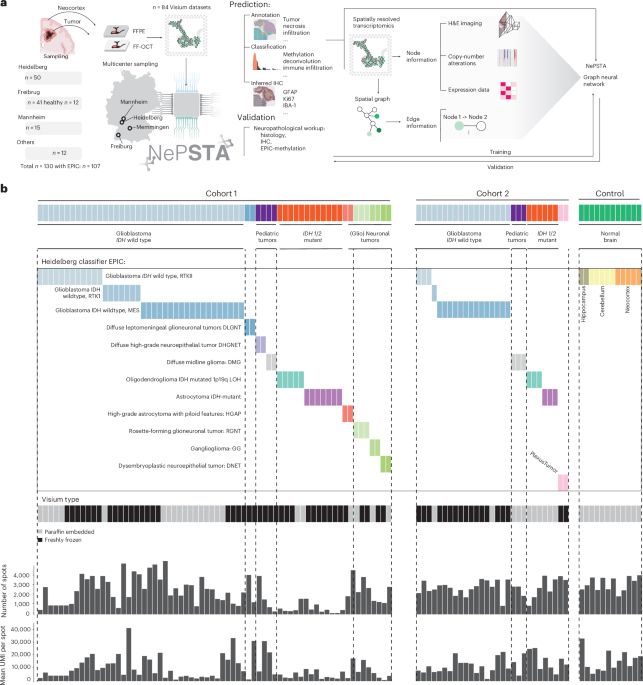
Spatially resolved transcriptomics and graph-based deep learning improve accuracy of routine CNS tumor diagnostics | @naturecancer.bsky.social
www.nature.com/articles/s43...
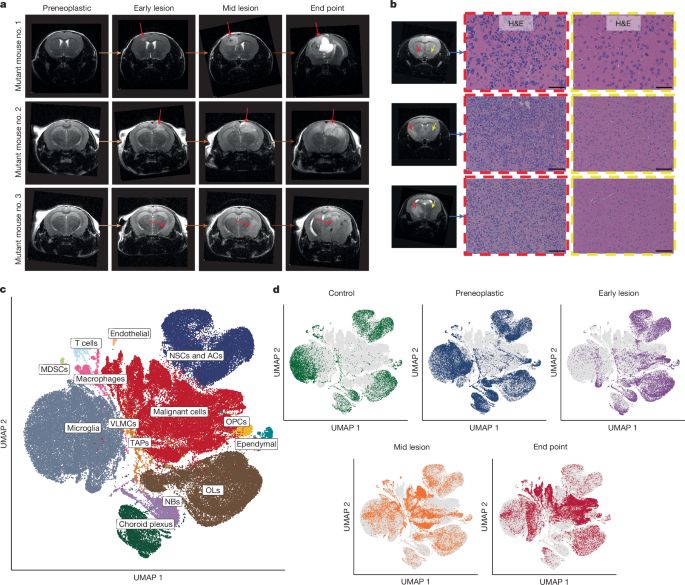
biorxiv.org/content/10.1101/2024.12.11.627935v1