
Genetics PhD @StanfordMed, BS @UArkansas
elsi / rna-seq / rare disease / multi-omics / chronic illness / x-chromosome



🧬 www.nature.com/articles/s41...

🧬 www.nature.com/articles/s41...
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
Having an extra sex chromosome really challenge the binary definition of sex based on XX and XY. So it is not only a medical, but also an important societal question.
📄 #PheWAS of male and female sex chromosome trisomies in 1.5 million participants of MVP, FinnGen, and UK Biobank
@finngen.bsky.social @ukbiobank.bsky.social

Having an extra sex chromosome really challenge the binary definition of sex based on XX and XY. So it is not only a medical, but also an important societal question.








I am eternally grateful for my editor, who took the original draft and helped me narrow down my key points into a more effective piece with a broader reach.
Also, thanks to the editorial & legal teams, who maintained the message while protecting us in this fraught moment.
https://go.nature.com/4jGqiUc

I am eternally grateful for my editor, who took the original draft and helped me narrow down my key points into a more effective piece with a broader reach.
Also, thanks to the editorial & legal teams, who maintained the message while protecting us in this fraught moment.
Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...




Check out SCOPE! An org @vangeliqueallen.bsky.social and I created! First panel on creating children’s books and podcasts is today at 1pm ET 😊🧪

Check out SCOPE! An org @vangeliqueallen.bsky.social and I created! First panel on creating children’s books and podcasts is today at 1pm ET 😊🧪

www.scientificamerican.com/article/beyo...
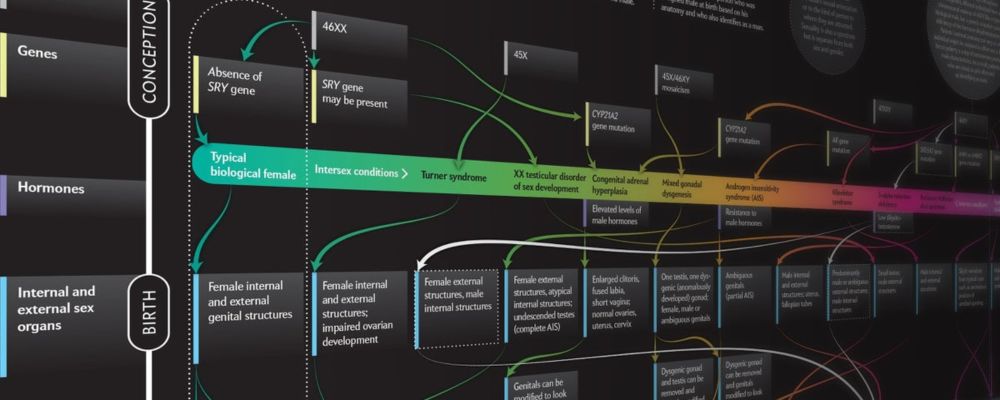
www.scientificamerican.com/article/beyo...
A transcriptome first approach identifies pathogenic variants in RNU4ATAC and highlights the RNU6ATAC as a putative new disease gene 🧬🖥️
A 💪 day for snRNA preprints!!

A transcriptome first approach identifies pathogenic variants in RNU4ATAC and highlights the RNU6ATAC as a putative new disease gene 🧬🖥️
A 💪 day for snRNA preprints!!

Accelerating #RareDisease diagnostics with cutting-edge #Genomics and global data sharing of omics and deep phenotyping from ~7500 individuals on NHGRI AnVIL and much more to come! 🧬

Accelerating #RareDisease diagnostics with cutting-edge #Genomics and global data sharing of omics and deep phenotyping from ~7500 individuals on NHGRI AnVIL and much more to come! 🧬
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
Previous tweetorial: x.com/MoezDawood/s...
Previous post by @ee-reh-neh.bsky.social: bsky.app/profile/ee-r...
Previous tweetorial: x.com/MoezDawood/s...
Previous post by @ee-reh-neh.bsky.social: bsky.app/profile/ee-r...

