Today we’re announcing global democratization of deidentified allele count + frequency data with population breakdown from the first ~250k short-read WGS in All of Us designed to plug straight into clinical workflows. It is ~1.1 billion unique variants! 🧬💡
🧵 (1/4)

#opensource

#opensource
1/9
1/9
arxiv.org/abs/2503.18810

arxiv.org/abs/2503.18810
It was a pleasure to work with @calhoujd.bsky.social and all the coauthors brought together by @varianteffect.bsky.social.
arxiv.org/abs/2503.18810

It was a pleasure to work with @calhoujd.bsky.social and all the coauthors brought together by @varianteffect.bsky.social.
Today we’re announcing global democratization of deidentified allele count + frequency data with population breakdown from the first ~250k short-read WGS in All of Us designed to plug straight into clinical workflows. It is ~1.1 billion unique variants! 🧬💡
🧵 (1/4)
Today we’re announcing global democratization of deidentified allele count + frequency data with population breakdown from the first ~250k short-read WGS in All of Us designed to plug straight into clinical workflows. It is ~1.1 billion unique variants! 🧬💡
🧵 (1/4)
Don't miss @moezdawood.bsky.social presenting "GREGoR: Accelerating Genomics for Rare Diseases"
Friday 3/20 at 1:30pm PST
Platform Session 8
Preprint here: pubmed.ncbi.nlm.nih.gov/39764392/
#Genomics #Research #Collaboration @gregor-research.bsky.social

Don't miss @moezdawood.bsky.social presenting "GREGoR: Accelerating Genomics for Rare Diseases"
Friday 3/20 at 1:30pm PST
Platform Session 8
Preprint here: pubmed.ncbi.nlm.nih.gov/39764392/
#Genomics #Research #Collaboration @gregor-research.bsky.social
doi.org/10.1101/2025...
1/n
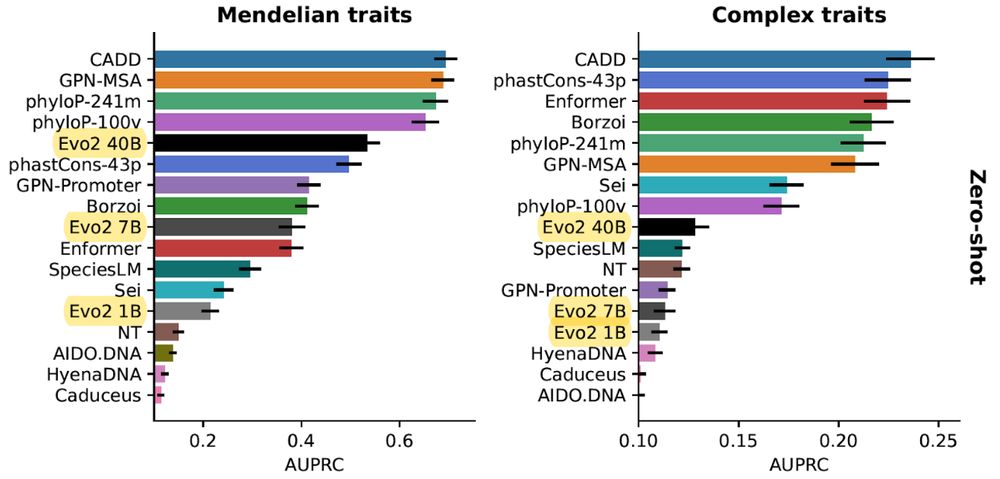
doi.org/10.1101/2025...
1/n
preprint: www.medrxiv.org/content/10.1...
R package: github.com/sbergercnmc/...
(1/n)
preprint: www.medrxiv.org/content/10.1...
R package: github.com/sbergercnmc/...
(1/n)
Just in time for #RareDisease Day, the @gene2phenotype.bsky.social website has a fresh look & new features to improve access to gene-disease models. 🧬💻
👉 Updated website: www.ebi.ac.uk/gene2phenoty...
👉 Why it matters: www.ebi.ac.uk/about/news/u...
Available at www.ebi.ac.uk/gene2phenotype.
Just in time for #RareDisease Day, the @gene2phenotype.bsky.social website has a fresh look & new features to improve access to gene-disease models. 🧬💻
👉 Updated website: www.ebi.ac.uk/gene2phenoty...
👉 Why it matters: www.ebi.ac.uk/about/news/u...

Rubisco is the main CO2-fixing enzyme of the biosphere yet its kinetics are slow. This work shows that non-trivial biochemical changes are readily accessible pointing the way to new engineering ways
🧪 @nature.com
www.nature.com/articles/s41...

Rubisco is the main CO2-fixing enzyme of the biosphere yet its kinetics are slow. This work shows that non-trivial biochemical changes are readily accessible pointing the way to new engineering ways
🧪 @nature.com
www.nature.com/articles/s41...
arxiv.org/abs/2501.04822

arxiv.org/abs/2501.04822

Accelerating #RareDisease diagnostics with cutting-edge #Genomics and global data sharing of omics and deep phenotyping from ~7500 individuals on NHGRI AnVIL and much more to come! 🧬

Accelerating #RareDisease diagnostics with cutting-edge #Genomics and global data sharing of omics and deep phenotyping from ~7500 individuals on NHGRI AnVIL and much more to come! 🧬

Accelerating #RareDisease diagnostics with cutting-edge #Genomics and global data sharing of omics and deep phenotyping from ~7500 individuals on NHGRI AnVIL and much more to come! 🧬

Previous tweetorial: x.com/MoezDawood/s...
Previous post by @ee-reh-neh.bsky.social: bsky.app/profile/ee-r...
Previous tweetorial: x.com/MoezDawood/s...
Previous post by @ee-reh-neh.bsky.social: bsky.app/profile/ee-r...


