Sporadic on social media.
🧬 www.nature.com/articles/s41...

🧬 www.nature.com/articles/s41...

(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com

(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com
#RareDisease #Research

#RareDisease #Research
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
💥Full list & details: www.ashg.org/membership/a...
#ASHG #HumanGenetics

💥Full list & details: www.ashg.org/membership/a...
#ASHG #HumanGenetics
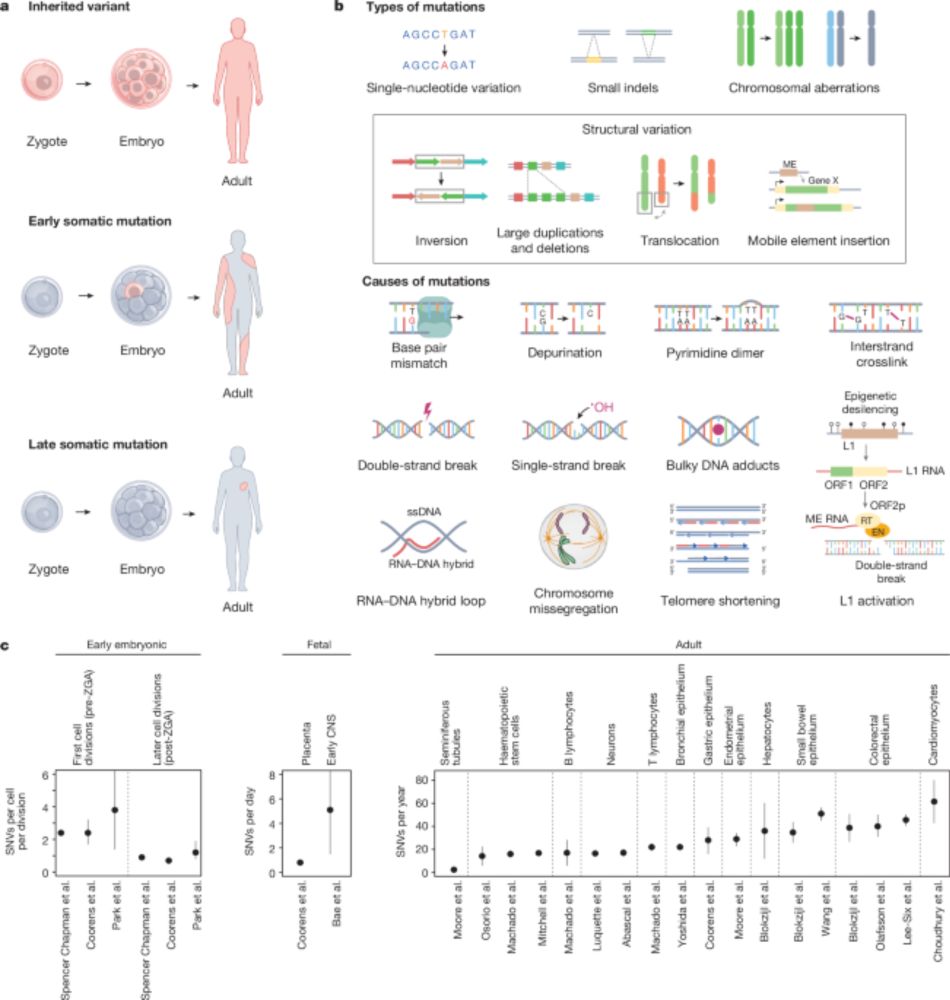
(For the record, no one has ever asked me this, but it is a really good question!)
I think we know where they are.
(For the record, no one has ever asked me this, but it is a really good question!)
I think we know where they are.


complex trait-associated variants” Very interesting preprint by Kathryn Lawrence @tamigj.bsky.social @sbmontgom.bsky.social
good arguments to move beyond single-gene-at-a-time approaches 🧪🧬
www.biorxiv.org/content/10.1...

complex trait-associated variants” Very interesting preprint by Kathryn Lawrence @tamigj.bsky.social @sbmontgom.bsky.social
good arguments to move beyond single-gene-at-a-time approaches 🧪🧬
www.biorxiv.org/content/10.1...
📄Transcriptomic signatures of rare variant impacts across sex and the X-chromosome
🧑🤝🧑 @raungar.bsky.social @sbmontgom.bsky.social & co
👉https://bit.ly/4kmWc8Z

📄Transcriptomic signatures of rare variant impacts across sex and the X-chromosome
🧑🤝🧑 @raungar.bsky.social @sbmontgom.bsky.social & co
👉https://bit.ly/4kmWc8Z
Building AI models or the data to train them?
Core funding of >$130M a year for a faculty of ~30.
www.nature.com/naturecareer...
acrobat.adobe.com/id/urn:aaid:...
pls RT!

Building AI models or the data to train them?
Core funding of >$130M a year for a faculty of ~30.
www.nature.com/naturecareer...
acrobat.adobe.com/id/urn:aaid:...
pls RT!
www.biorxiv.org/content/10.1...
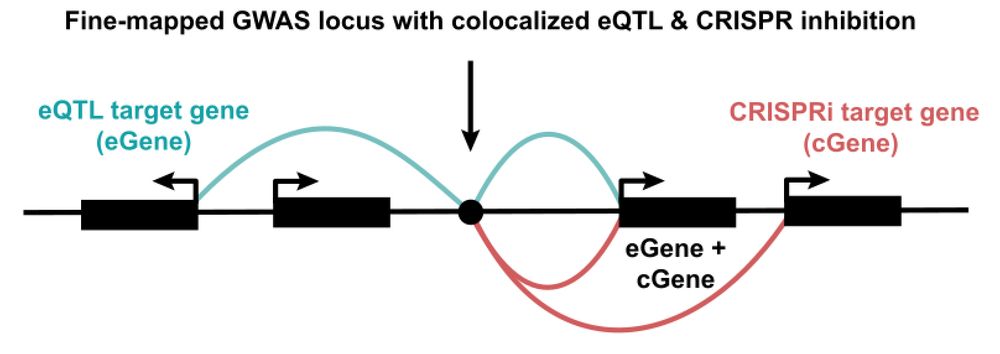
www.biorxiv.org/content/10.1...


academic.oup.com/nar/article/...

academic.oup.com/nar/article/...

📅 Dates: 9-11 April 2025
💭 Share insights in person
Explore the latest #genomics advances accelerating improvements in clinical care for rare disorders, globally.
⏰Secure your place by 11 March: bit.ly/3BpAe44

📅 Dates: 9-11 April 2025
💭 Share insights in person
Explore the latest #genomics advances accelerating improvements in clinical care for rare disorders, globally.
⏰Secure your place by 11 March: bit.ly/3BpAe44

📅 Dates: 9-11 April 2025
💭 Share insights in person
Explore the latest #genomics advances accelerating improvements in clinical care for rare disorders, globally.
⏰Secure your place by 11 March: bit.ly/3BpAe44
Our new paper, led by the outstanding Maxim Zaslavsky @maximzaslavsky.bsky.social sky.bsky.social with help from me and Anshul Kundaje @anshulkundaje.bsky.social.
Link: buff.ly/3QvxSVf
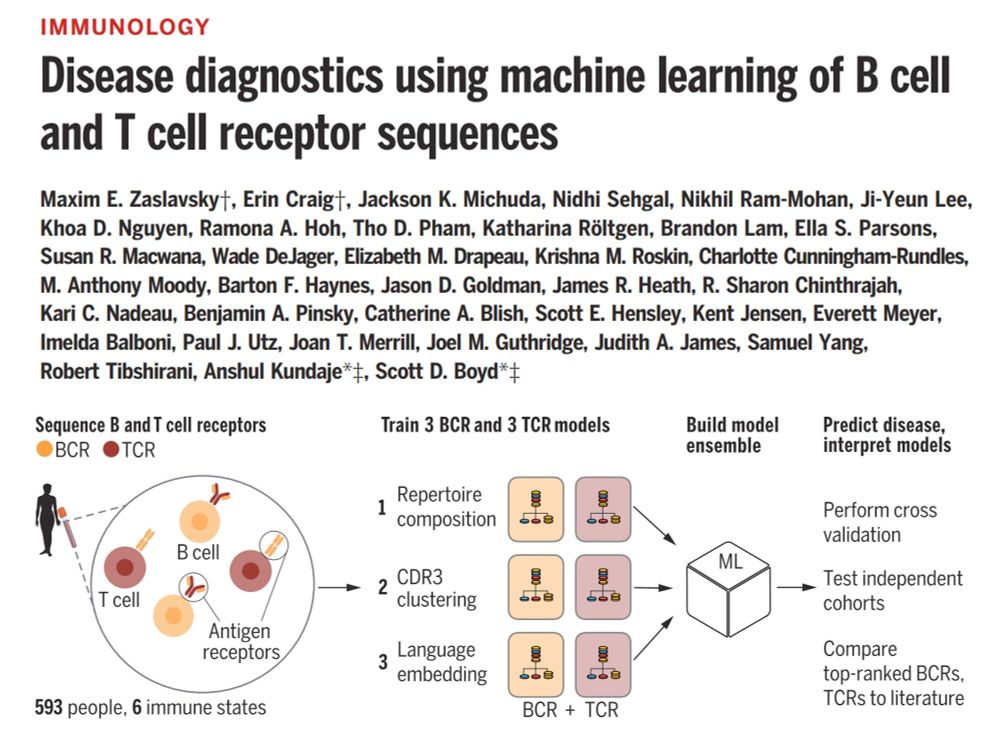
Our new paper, led by the outstanding Maxim Zaslavsky @maximzaslavsky.bsky.social sky.bsky.social with help from me and Anshul Kundaje @anshulkundaje.bsky.social.
Link: buff.ly/3QvxSVf
Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

