
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
To understand gene regulation across diverse environmental conditions and cellular contexts, we treated a broad array of human cell types with three environmental exposures in vitro.
www.biorxiv.org/content/10.1...
To understand gene regulation across diverse environmental conditions and cellular contexts, we treated a broad array of human cell types with three environmental exposures in vitro.
www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
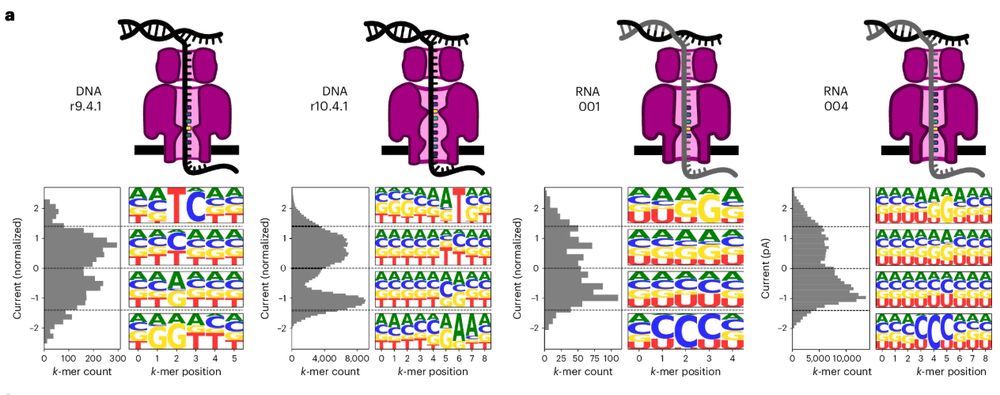
www.nature.com/articles/s41...


