📄 www.biorxiv.org/content/10.6...
📦 github.com/marbl/anianns

📄 www.biorxiv.org/content/10.6...
📦 github.com/marbl/anianns
employment.utah.edu/salt-lake-ci...
employment.utah.edu/salt-lake-ci...

It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025



It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025
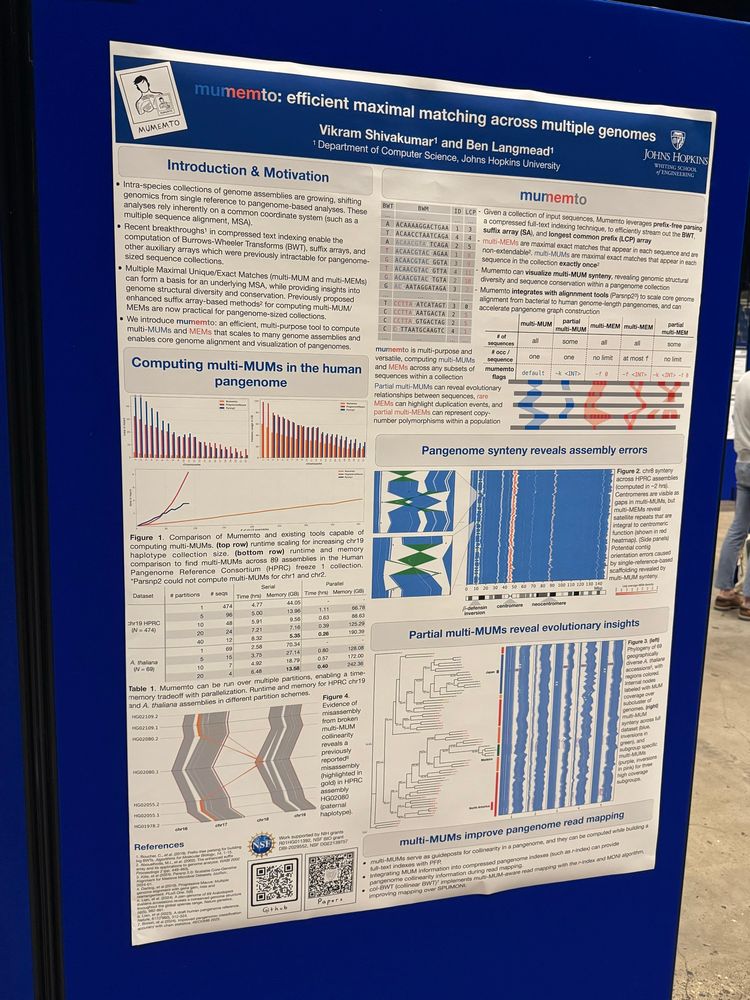
Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
Very efficient pangenome visualization tool, revealing synteny and variations!



Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
Very efficient pangenome visualization tool, revealing synteny and variations!


Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)

Computing average nucleotide identity (ANI) is neither conceptually nor computationally trivial. Its definition has evolved over years, with different meanings and assumptions (1/5)
Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025



Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025

*maximal unique matches in pangenomes, now if you did that on sequenced moms you could do mummoms

*maximal unique matches in pangenomes, now if you did that on sequenced moms you could do mummoms
github.com/mohsenzakeri...
github.com/mohsenzakeri...


