www.mohsenzakeri.com
List of talks and posters: meetings.cshl.edu/abstracts.as...

List of talks and posters: meetings.cshl.edu/abstracts.as...






If you're a PI working in algorithmic genomics (& you can recommend my lab to your top graduating students ;P), please let them know!
An great teamwork by @mohsenzakeri.bsky.social, @stephenhwang.bsky.social and me, with the excellent mentorship of @benlangmead.bsky.social
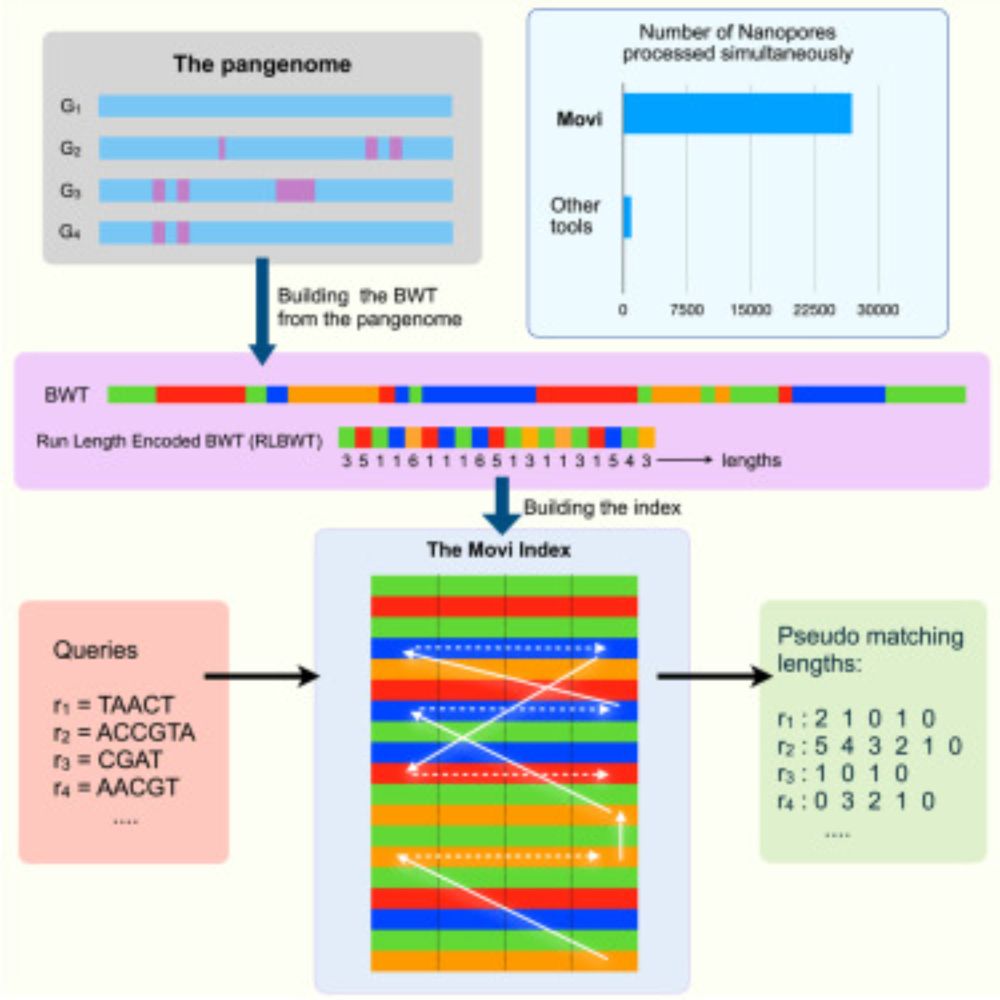
An great teamwork by @mohsenzakeri.bsky.social, @stephenhwang.bsky.social and me, with the excellent mentorship of @benlangmead.bsky.social
Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025



Github: github.com/vikshiv/mume...
First paper: genomebiology.biomedcentral.com/articles/10....
Second: www.biorxiv.org/content/10.1... #WABI2025
wabiconf.github.io/2025/talks/t...
wabiconf.github.io/2025/talks/t...
👉 Code & community: github.com/Kuanhao-Chao...
It’s been incredibly rewarding building this for the genomics community. Can’t wait for your feedback and contributions!
👉 Code & community: github.com/Kuanhao-Chao...
It’s been incredibly rewarding building this for the genomics community. Can’t wait for your feedback and contributions!
github.com/mohsenzakeri...
github.com/mohsenzakeri...
github.com/mohsenzakeri...
Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n

Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n
* abstract deadline: May 12 (AoE)
* paper deadline: May 15 (AoE)
Consider submitting your exciting algorithmic bioinformatics work to the WABI conference!
* abstract deadline: May 12 (AoE)
* paper deadline: May 15 (AoE)
Consider submitting your exciting algorithmic bioinformatics work to the WABI conference!
You can find out more about me at these links:
LinkedIn: www.linkedin.com/in/arun96/
Personal Website: arundas.org
You can find out more about me at these links:
LinkedIn: www.linkedin.com/in/arun96/
Personal Website: arundas.org
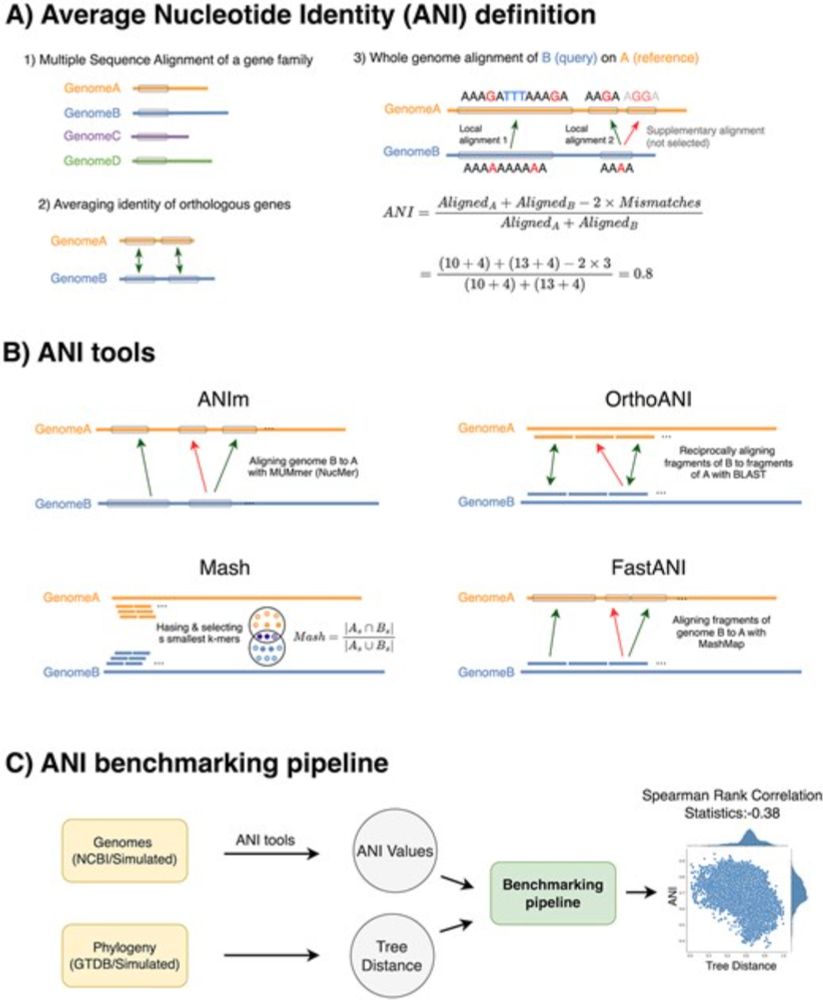
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
If you want to learn more about tree-based quantification & differential testing, or scATAC-seq preprocessing; tune in!
Talk link: umd.zoom.us/j/9873133564...
Abstract: talks.cs.umd.edu/talks/4137
If you want to learn more about tree-based quantification & differential testing, or scATAC-seq preprocessing; tune in!
Talk link: umd.zoom.us/j/9873133564...
Abstract: talks.cs.umd.edu/talks/4137
🌟 Alicia Oshlack – computational transcriptomics
@aliciao.bsky.social
🌟 Rayan Chikhi – sequencing data structures
@rayanchikhi.bsky.social
🗓️ Dates: April 24–25, 2025
📍 Seoul, South Korea
recomb-seq.github.io/speakers/


🌟 Alicia Oshlack – computational transcriptomics
@aliciao.bsky.social
🌟 Rayan Chikhi – sequencing data structures
@rayanchikhi.bsky.social
🗓️ Dates: April 24–25, 2025
📍 Seoul, South Korea
recomb-seq.github.io/speakers/