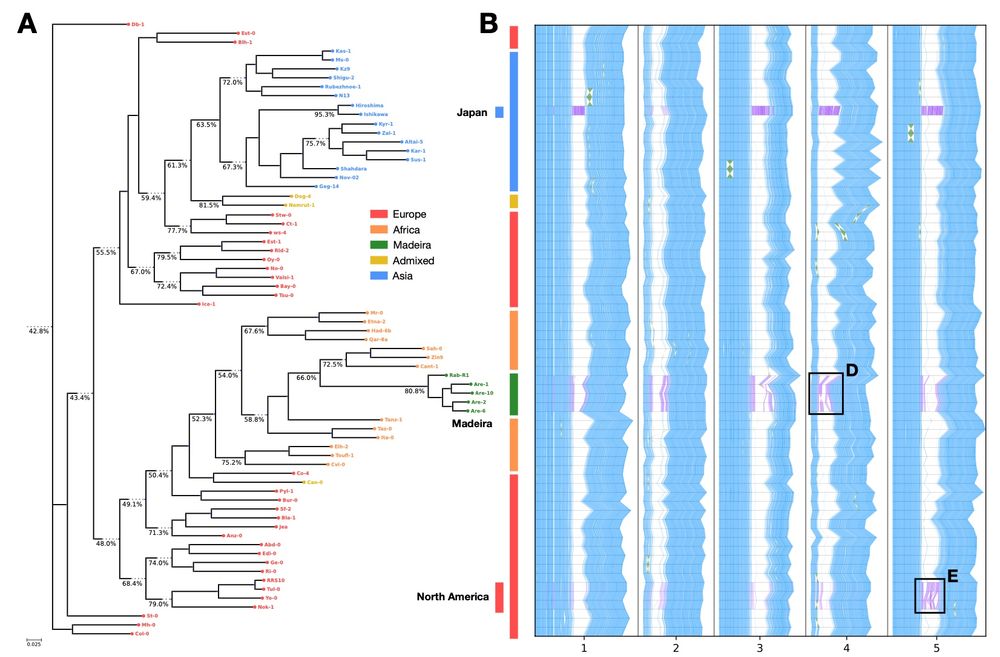
It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025



It uses k-mer conservation to annotate genomic variation across hundreds of genomes, followed by normalization of k-mer profiles to identify introgression events
github.com/kjenike/pana... #GI2025
Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
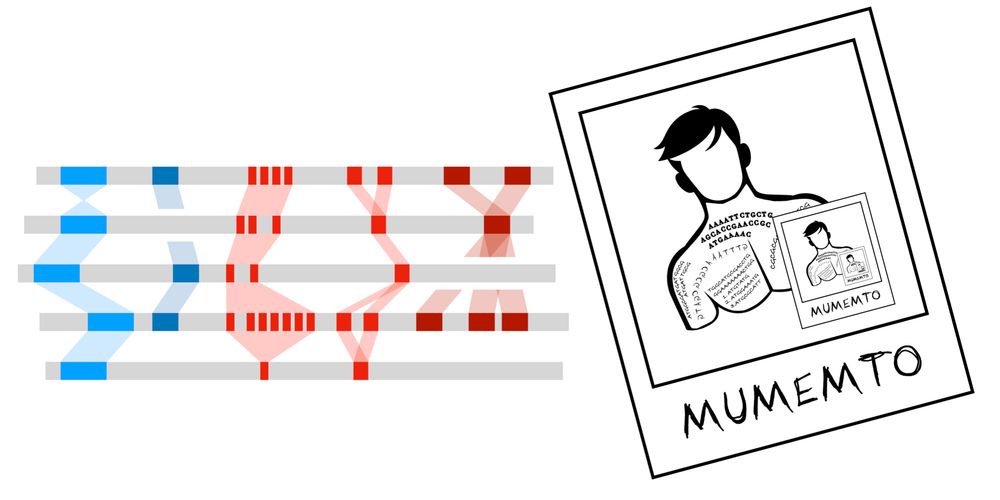
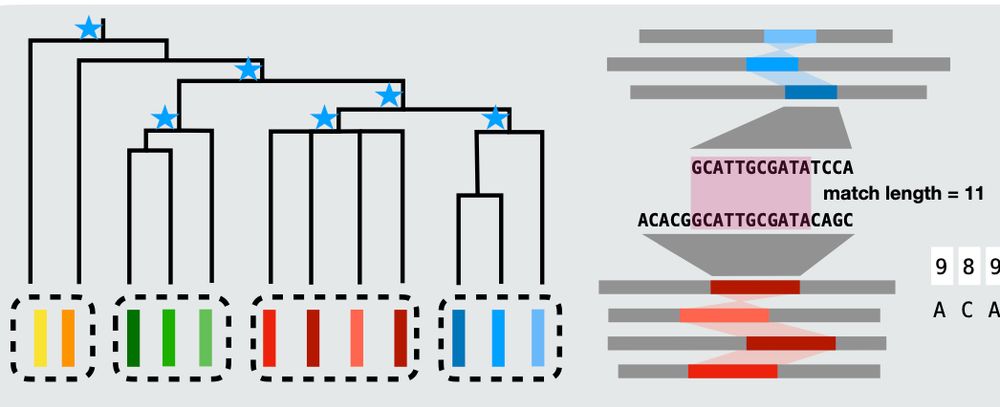
Very efficient pangenome visualization tool, revealing synteny and variations!



Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
Very efficient pangenome visualization tool, revealing synteny and variations!