ORCID: 0000-0002-1143-5961
✴️ genetic drug design
✴️ the Big Bang's first microseconds
✴️ crowd movement
✴️ nature-society interactions
👉 buff.ly/ZQhLp4h
Congrats to the 66 awarded research teams!
#FrontierResearch



@dunhamlab.bsky.social and #HouLabTJU.
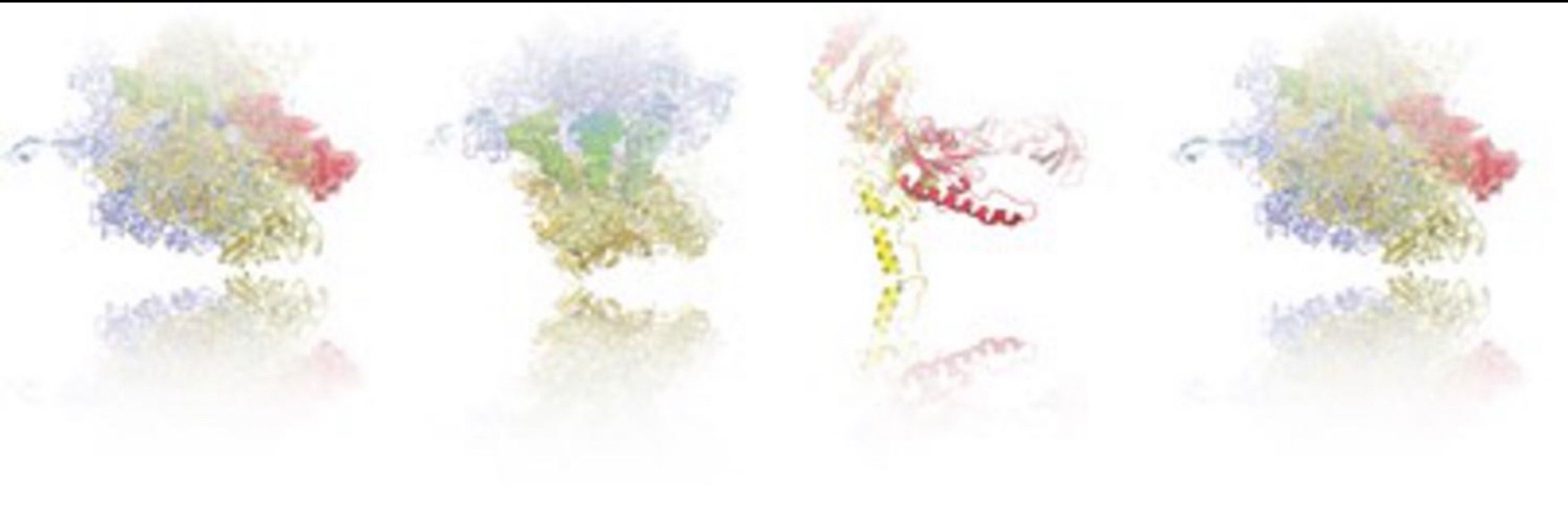
In this Nat Commun paper, we combine smFRET and cryo-EM to show how the tRNA modification m¹G37 stabilizes the reading frame—and what happens when it’s missing.

@dunhamlab.bsky.social and #HouLabTJU.
In this Nat Commun paper, we combine smFRET and cryo-EM to show how the tRNA modification m¹G37 stabilizes the reading frame—and what happens when it’s missing.

thanks to @i2bcparissaclay.bsky.social for its financial support through epiRNA programs
academic.oup.com/nargab/artic...

thanks to @i2bcparissaclay.bsky.social for its financial support through epiRNA programs
academic.oup.com/nargab/artic...
Hypoxia-induced ribosomal RNA modifications in the peptidyl-transferase center contribute to anaerobic growth of bacteria: Molecular Cell www.cell.com/molecular-ce...

Hypoxia-induced ribosomal RNA modifications in the peptidyl-transferase center contribute to anaerobic growth of bacteria: Molecular Cell www.cell.com/molecular-ce...
Upstream start codons can produce alternative proteins with novel N-terminal amino acids that block signal peptides.
These proteins are translated but not found in any proteomics experiments.
rdcu.be/eUi7o
Upstream start codons can produce alternative proteins with novel N-terminal amino acids that block signal peptides.
These proteins are translated but not found in any proteomics experiments.
rdcu.be/eUi7o
Beyond RNA modification: a tRNA-modifying enzyme shaping oxidative stress resilience and metabolism in V. cholerae #rnasky #rnabiology
😊🦠💫

Beyond RNA modification: a tRNA-modifying enzyme shaping oxidative stress resilience and metabolism in V. cholerae #rnasky #rnabiology
😊🦠💫



They'll investigate premature termination codons, the genetic 'stop signals' that cut production of proteins short causing disorders. 🧬🔬
👉 buff.ly/ZQhLp4h
#ERCSyG

They'll investigate premature termination codons, the genetic 'stop signals' that cut production of proteins short causing disorders. 🧬🔬
👉 buff.ly/ZQhLp4h
#ERCSyG
✴️ genetic drug design
✴️ the Big Bang's first microseconds
✴️ crowd movement
✴️ nature-society interactions
👉 buff.ly/ZQhLp4h
Congrats to the 66 awarded research teams!
#FrontierResearch

Bacteria rewire their RNA world under stress & infection. tRNA mods as dual-function sensors, phage tRNA slashers, hibernating ribosomes & haunted translation hubs.
🧬🧪 Full November 1st issue → #RNAmod #Epitranscriptomics #HalloweenScience
#rnasky #microsky

Bacteria rewire their RNA world under stress & infection. tRNA mods as dual-function sensors, phage tRNA slashers, hibernating ribosomes & haunted translation hubs.
🧬🧪 Full November 1st issue → #RNAmod #Epitranscriptomics #HalloweenScience
#rnasky #microsky

🧵 Thread below
Full analysis: https://helixbrief.com/article/a2e1736b-7031-4cdb-98d7-78309181890c
🧵 Thread below
Full analysis: https://helixbrief.com/article/a2e1736b-7031-4cdb-98d7-78309181890c
www.uni-hamburg.de/en/stellenan...



In @natmethods.nature.com we propose using "Translon" for any region decoded by the ribosome

www.nature.com/articles/s41...

www.nature.com/articles/s41...



