Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad

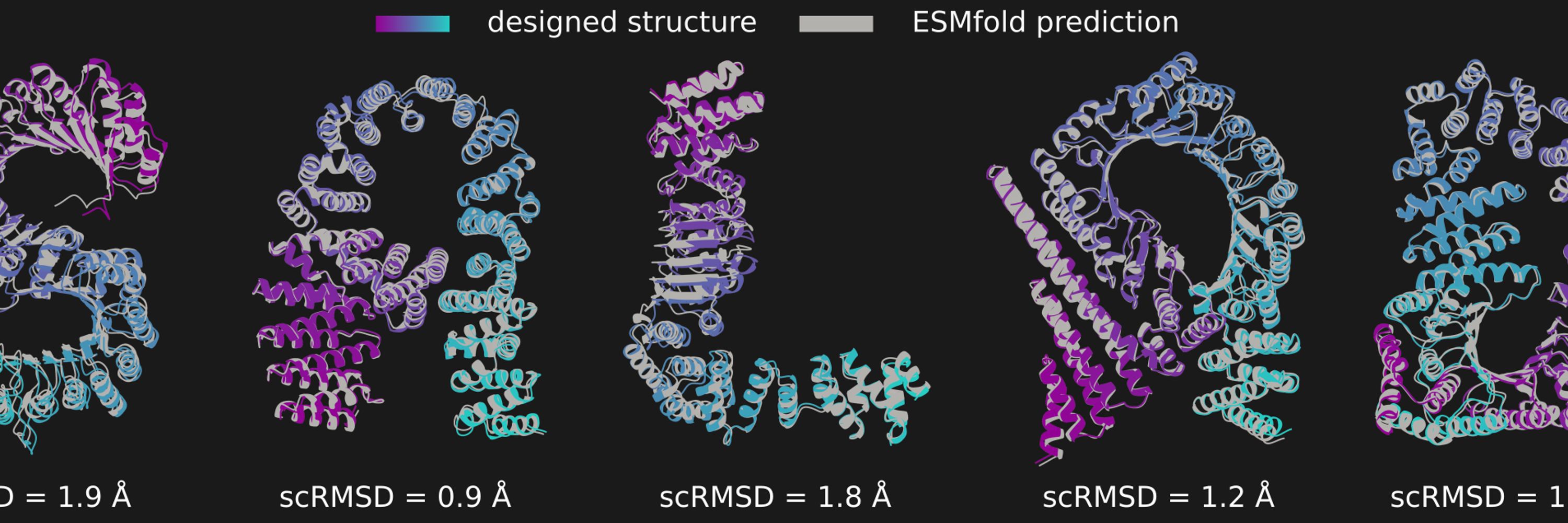
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)


www.cell.com/molecular-ce...

www.cell.com/molecular-ce...
mirdita.org

mirdita.org
doi.org/10.64898/202...

doi.org/10.64898/202...
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
doi.org/10.1002/pro....

doi.org/10.1002/pro....
One structure for each user with finalized structure from current year.
#crystallography #chemtree #chemchat #chemsky


One structure for each user with finalized structure from current year.
#crystallography #chemtree #chemchat #chemsky
Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...



Why not try bluesky-less Frosina's new self-supervised algorithm for semantic segmentation and particle picking for #teamtomo (includes a new denoiser without the need for odd/even tomos)
Code is on GitHub if you want to try it out

Why not try bluesky-less Frosina's new self-supervised algorithm for semantic segmentation and particle picking for #teamtomo (includes a new denoiser without the need for odd/even tomos)
Code is on GitHub if you want to try it out
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
cryoJAX: A Cryo-EM Image Simulation Library In JAX
www.biorxiv.org/content/10.1...
cryoJAX is a modular framework for implementing forward models of cryo-EM image formation. these can be used to build powerful data analyses!
cryoJAX: A Cryo-EM Image Simulation Library In JAX
www.biorxiv.org/content/10.1...
cryoJAX is a modular framework for implementing forward models of cryo-EM image formation. these can be used to build powerful data analyses!
(Please repost to reach a broad audience.)
(Please repost to reach a broad audience.)
100 designs
⬇️
34 got through the screening (ipTM, PAE, RMSD)
⬇️
4 ordered (gene synthesis + codon optimization)
⬇️
3 expressed and soluble
⬇️
1 interacts with its target !!!!
Now the story begins !

100 designs
⬇️
34 got through the screening (ipTM, PAE, RMSD)
⬇️
4 ordered (gene synthesis + codon optimization)
⬇️
3 expressed and soluble
⬇️
1 interacts with its target !!!!
Now the story begins !
Researchers from EMBL's Korbel Group have developed a new AI method – MAGIC – which, through a game of molecular laser tag, is shedding light on how chromosomal abnormalities form in cells.
www.embl.org/news/science...

Researchers from EMBL's Korbel Group have developed a new AI method – MAGIC – which, through a game of molecular laser tag, is shedding light on how chromosomal abnormalities form in cells.
www.embl.org/news/science...
Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)

Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem


ista.ac.at/en/job/tenur...

ista.ac.at/en/job/tenur...


