Michael Jendrusch
@mjendrusch.bsky.social
(he/him) Former PhD student / Postdoc @ Korbel group, EMBL.
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Also, shoutout to @blender.org and @bradyajohnston.bsky.social for making it so easy to turn protein structures into nice images!
5/5
5/5
September 24, 2025 at 12:27 PM
Also, shoutout to @blender.org and @bradyajohnston.bsky.social for making it so easy to turn protein structures into nice images!
5/5
5/5
Compared to the preprint, we have added comparisons to Proteína (openreview.net/forum?id=TVQLu34…) in terms of unconditional design. Here, salad compares favorably in terms of both runtime per design and designability of generated structures.
3/🧵
3/🧵


September 24, 2025 at 12:27 PM
Compared to the preprint, we have added comparisons to Proteína (openreview.net/forum?id=TVQLu34…) in terms of unconditional design. Here, salad compares favorably in terms of both runtime per design and designability of generated structures.
3/🧵
3/🧵
Some salad news: The code (github.com/mjendrusch/salad) is now somewhat cleaned up and there is a Colab notebook for running a full salad -> ProteinMPNN -> AlphaFold2 pipeline to design proteins like those in the image:
colab.research.google.com/github/mjend...
More improvements coming soon!
colab.research.google.com/github/mjend...
More improvements coming soon!

May 21, 2025 at 3:40 PM
Some salad news: The code (github.com/mjendrusch/salad) is now somewhat cleaned up and there is a Colab notebook for running a full salad -> ProteinMPNN -> AlphaFold2 pipeline to design proteins like those in the image:
colab.research.google.com/github/mjend...
More improvements coming soon!
colab.research.google.com/github/mjend...
More improvements coming soon!
This tendency is also the case for failed designs (1,000 aa, failed, below). We did not include these in the data package, as the archive including everything was becoming uncomfortably large.

February 7, 2025 at 1:13 PM
This tendency is also the case for failed designs (1,000 aa, failed, below). We did not include these in the data package, as the archive including everything was becoming uncomfortably large.
You're right, denoising models seem to generate more idealised / boring structures, especially for larger proteins.
For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)
For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)

February 7, 2025 at 1:13 PM
You're right, denoising models seem to generate more idealised / boring structures, especially for larger proteins.
For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)
For smaller proteins, salad generations can get pretty crazy, though (400aa, upper row is the first 4 salad VP-scaled examples, lower row is the first 4 RSO examples I could find)
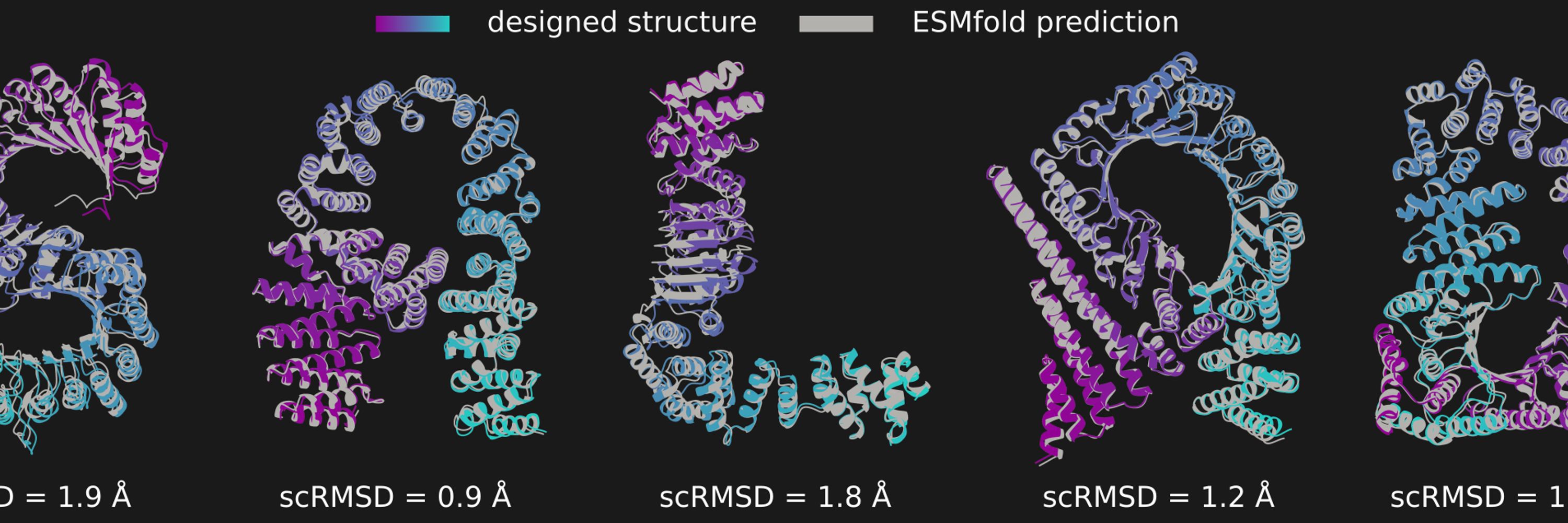
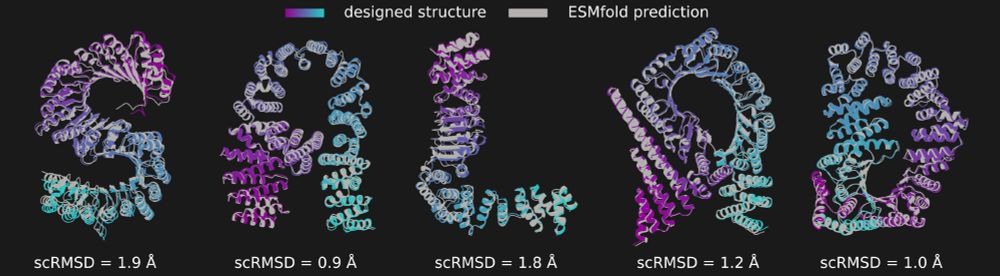
It also means that we can have salad spell out its name in proteins.
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
February 6, 2025 at 1:07 PM
It also means that we can have salad spell out its name in proteins.
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
In the image you can see salad-generated structures spelling the word "SALAD", overlaid with their ESMfold predictions and scRMSD.
(6/N)
... such as motif-scaffolding, designing screw-symmetric repeat proteins and multi-state protein design à la ProteinGenerator (cf. www.nature.com/articles/s41...).
(5/N)
(5/N)



February 6, 2025 at 1:07 PM
... such as motif-scaffolding, designing screw-symmetric repeat proteins and multi-state protein design à la ProteinGenerator (cf. www.nature.com/articles/s41...).
(5/N)
(5/N)
4. we can teach salad new tricks without having to re-train the model.
We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)
We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)

February 6, 2025 at 1:07 PM
4. we can teach salad new tricks without having to re-train the model.
We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)
We extend RFdiffusion's sampling trick for symmetry to arbitrary edits to the input and output of salad models at each generation step.
This way, we can make salad work on design tasks it was not trained for ...
(4/N)
3. salad models generate designable structures up to 1,000 amino acids long.
For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)
For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)

February 6, 2025 at 1:07 PM
3. salad models generate designable structures up to 1,000 amino acids long.
For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)
For proteins of this size one would otherwise have to use RSO or af2cycler so far – protein structure diffusion models do not work well.
Our best salad models (blue) bridge this designability gap to RSO.
(3/N)
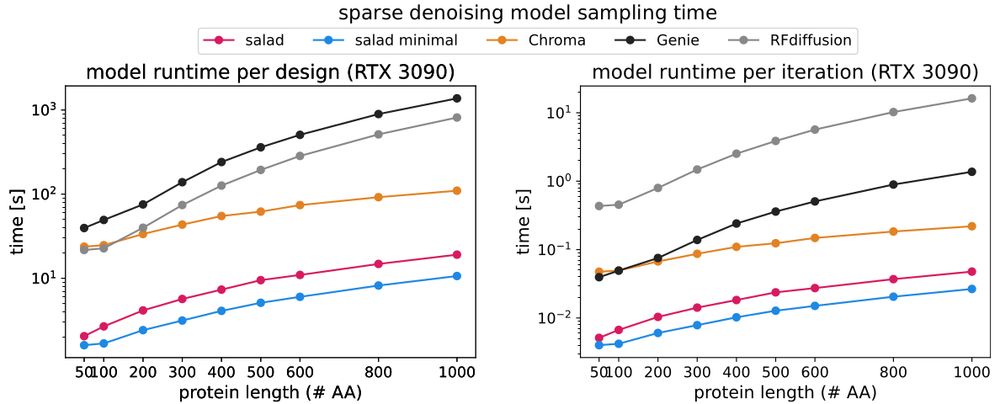
Why yet another protein structure generative model if we already have RFdiffusion?
(and Genie 2 and Chroma and Proteus and many others)
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
(and Genie 2 and Chroma and Proteus and many others)
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
February 6, 2025 at 1:07 PM
Why yet another protein structure generative model if we already have RFdiffusion?
(and Genie 2 and Chroma and Proteus and many others)
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
(and Genie 2 and Chroma and Proteus and many others)
1. salad is tiny, with the denoising model clocking in at about 8.4 M parameters.
2. salad is fast, up to 42x faster than RFdiffusion on large proteins
(2/N)
New protein ML preprint from my PhD project.
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)

February 6, 2025 at 1:07 PM
New protein ML preprint from my PhD project.
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)
Merry Christmas! May your protein backbones be designable, diverse and novel.
(now with 100% more structure predictions and plausibility metrics)
(now with 100% more structure predictions and plausibility metrics)

December 25, 2024 at 9:22 PM
Merry Christmas! May your protein backbones be designable, diverse and novel.
(now with 100% more structure predictions and plausibility metrics)
(now with 100% more structure predictions and plausibility metrics)
Reason Nr. 197 why working in protein design is fun:
you can spell out random messages as protein structures.
you can spell out random messages as protein structures.

December 24, 2024 at 3:02 PM
Reason Nr. 197 why working in protein design is fun:
you can spell out random messages as protein structures.
you can spell out random messages as protein structures.

