Michael Jendrusch
@mjendrusch.bsky.social
(he/him) Former PhD student / Postdoc @ Korbel group, EMBL.
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Also, shoutout to @blender.org and @bradyajohnston.bsky.social for making it so easy to turn protein structures into nice images!
5/5
5/5
September 24, 2025 at 12:27 PM
Also, shoutout to @blender.org and @bradyajohnston.bsky.social for making it so easy to turn protein structures into nice images!
5/5
5/5
While I was working on salad as part of my PhD at Korbel group at EMBL, it was very much a passion project. So it makes me very happy to see @embl.org feature this paper!
Thanks again to Jan and the team supporting me on my more non-standard projects!
4/🧵
Thanks again to Jan and the team supporting me on my more non-standard projects!
4/🧵
September 24, 2025 at 12:27 PM
While I was working on salad as part of my PhD at Korbel group at EMBL, it was very much a passion project. So it makes me very happy to see @embl.org feature this paper!
Thanks again to Jan and the team supporting me on my more non-standard projects!
4/🧵
Thanks again to Jan and the team supporting me on my more non-standard projects!
4/🧵
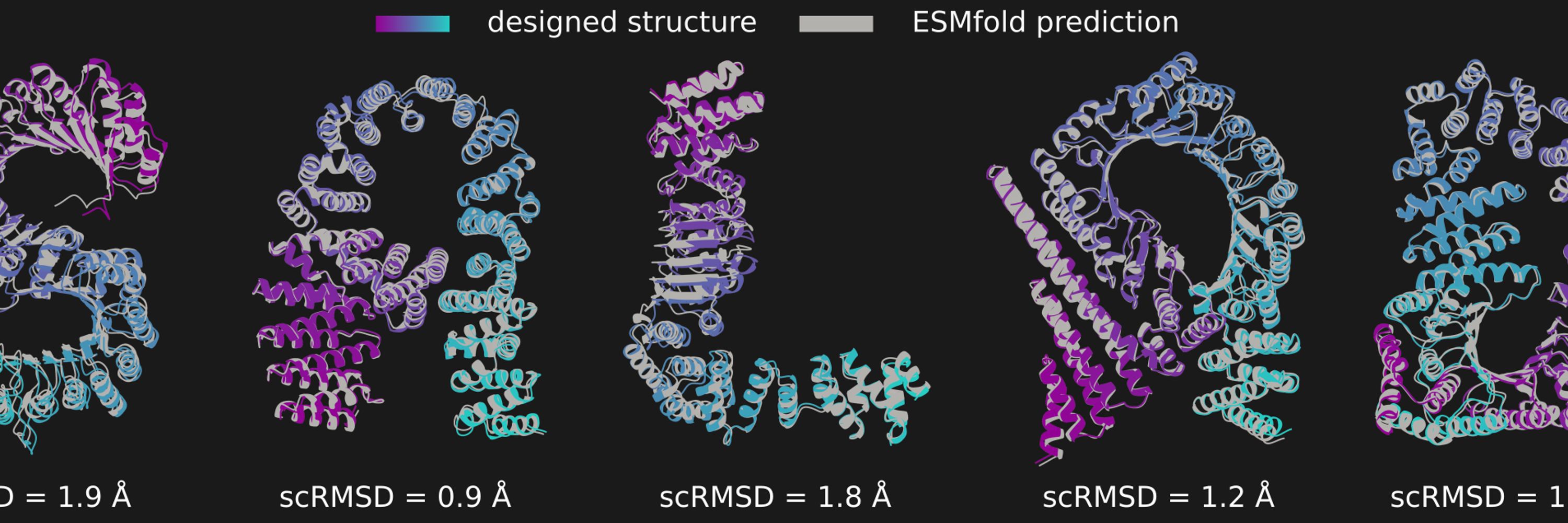
Compared to the preprint, we have added comparisons to Proteína (openreview.net/forum?id=TVQLu34…) in terms of unconditional design. Here, salad compares favorably in terms of both runtime per design and designability of generated structures.
3/🧵
3/🧵


September 24, 2025 at 12:27 PM
Compared to the preprint, we have added comparisons to Proteína (openreview.net/forum?id=TVQLu34…) in terms of unconditional design. Here, salad compares favorably in terms of both runtime per design and designability of generated structures.
3/🧵
3/🧵
The preprint and code have been out for a while - if you want a quick overview, have a look at the thread we did back in February:
bsky.app/profile/mjendrus…
If you want to try salad in your browser, you can load up this Colab notebook:
colab.research.google.com/github/mjendrusc…
2/🧵
bsky.app/profile/mjendrus…
If you want to try salad in your browser, you can load up this Colab notebook:
colab.research.google.com/github/mjendrusc…
2/🧵
New protein ML preprint from my PhD project.
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)
We describe salad (sparse all-atom denoising), a family of efficient protein structure diffusion models and show that it works well on a bunch of protein design task previously described in the literature.
Preprint: www.biorxiv.org/content/10.1...
(1/N)

September 24, 2025 at 12:27 PM
The preprint and code have been out for a while - if you want a quick overview, have a look at the thread we did back in February:
bsky.app/profile/mjendrus…
If you want to try salad in your browser, you can load up this Colab notebook:
colab.research.google.com/github/mjendrusc…
2/🧵
bsky.app/profile/mjendrus…
If you want to try salad in your browser, you can load up this Colab notebook:
colab.research.google.com/github/mjendrusc…
2/🧵

