Microbial evolution | Human microbiome ecology | Infectious disease epidemiology
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10

Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10

www.nature.com/articles/s41...

www.nature.com/articles/s41...
Excited to share this preprint in collaboration with @mboum.bsky.social @anaellefait.bsky.social Silvio Brugger and Alex Hall.
www.biorxiv.org/content/10.6...

Excited to share this preprint in collaboration with @mboum.bsky.social @anaellefait.bsky.social Silvio Brugger and Alex Hall.
www.biorxiv.org/content/10.6...

'Nationwide Clostridioides difficile population dynamics'
www.findaphd.com/phds/project...
Deadline for applications 9 Jan 2026, UK students only.

'Nationwide Clostridioides difficile population dynamics'
www.findaphd.com/phds/project...
Deadline for applications 9 Jan 2026, UK students only.
Huge congrats to @vhrcabral.bsky.social and @karinaxavierlab.bsky.social on this major work.
We found that our Klebsiella ARO112 can break the antibiotic/inflammation cycle in an IBD model

Huge congrats to @vhrcabral.bsky.social and @karinaxavierlab.bsky.social on this major work.
We found that our Klebsiella ARO112 can break the antibiotic/inflammation cycle in an IBD model

#phagesky
www.nature.com/articles/s41...

#phagesky
www.nature.com/articles/s41...
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8

Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8
We're looking for someone keen on bioinformatics and microbiome evolution.
Important info below on eligibility & URSA competition funding👇
www.findaphd.com/phds/project...

We're looking for someone keen on bioinformatics and microbiome evolution.
Important info below on eligibility & URSA competition funding👇
www.findaphd.com/phds/project...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
🧵 (1/14)
www.doi.org/10.1038/s414...
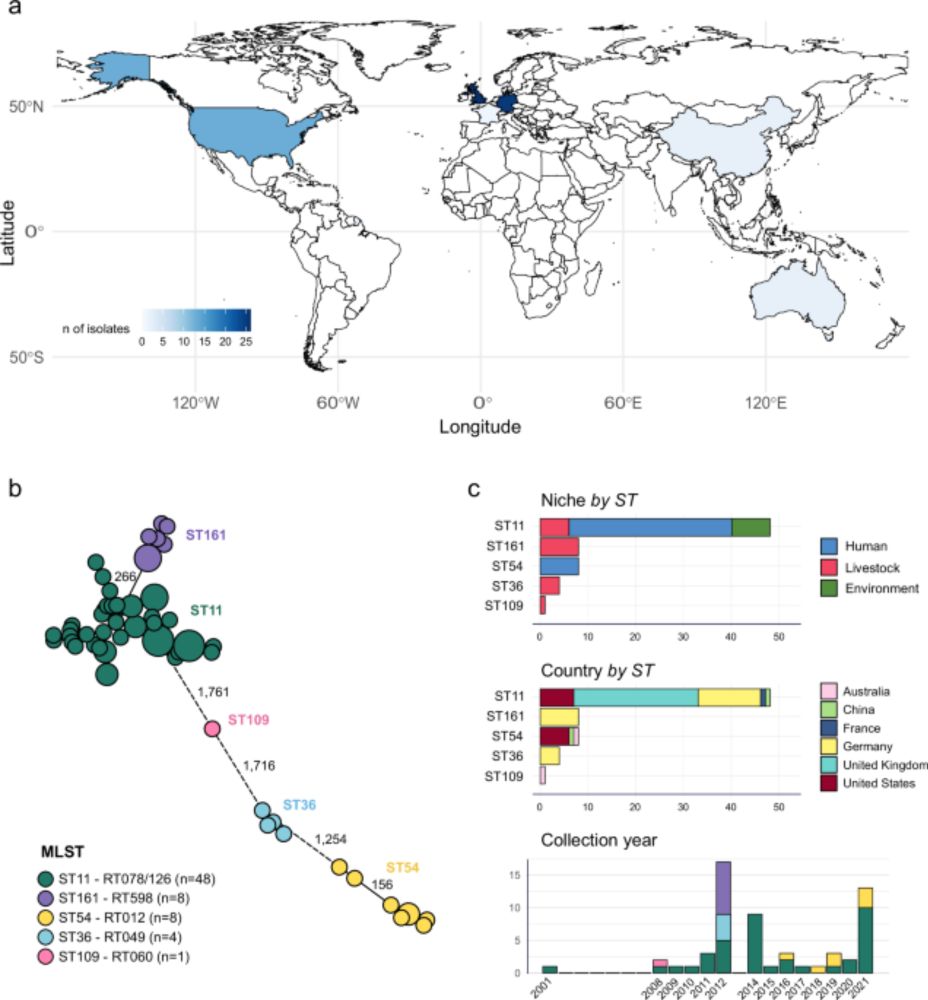
🧵 (1/14)
www.doi.org/10.1038/s414...

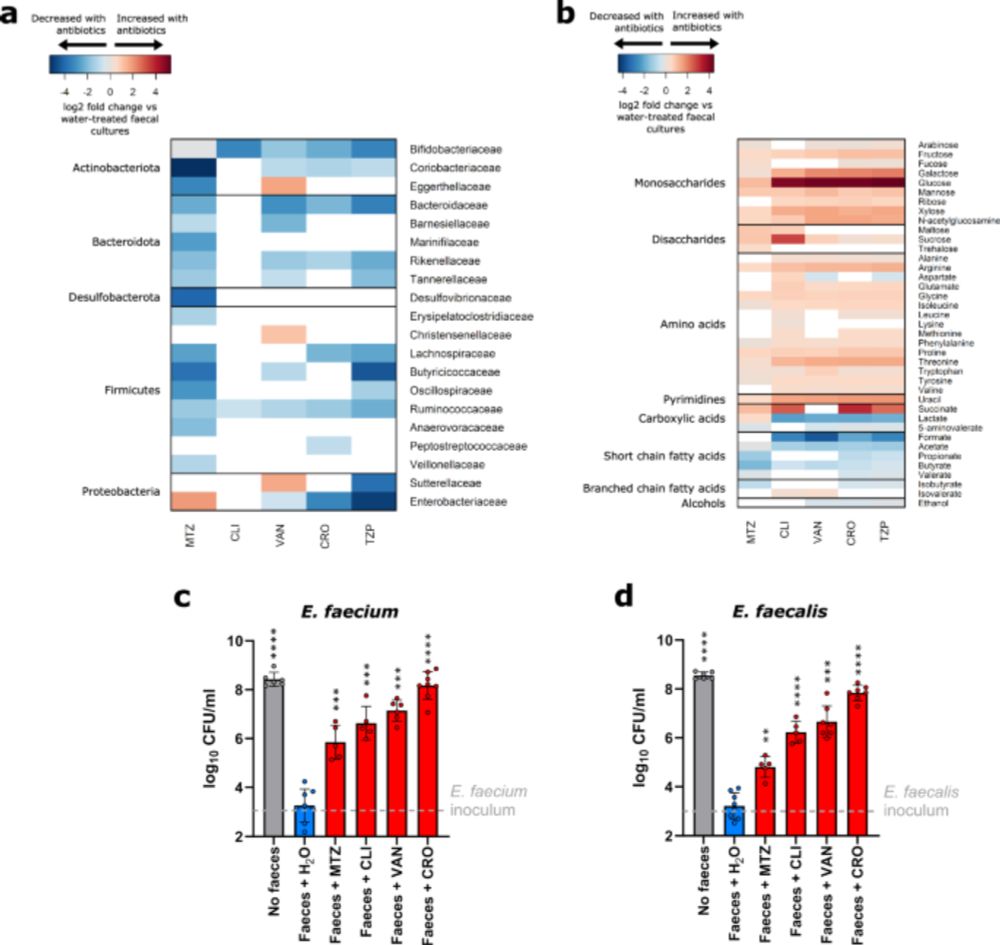
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63

www.nature.com/articles/s41...

www.nature.com/articles/s41...
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social



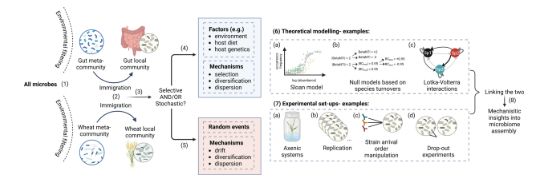
PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
A discussion on the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host genetic diversity, and the implications for transmission analysis and infectious disease control.
#MicroSky 🦠
www.nature.com/articles/s41...
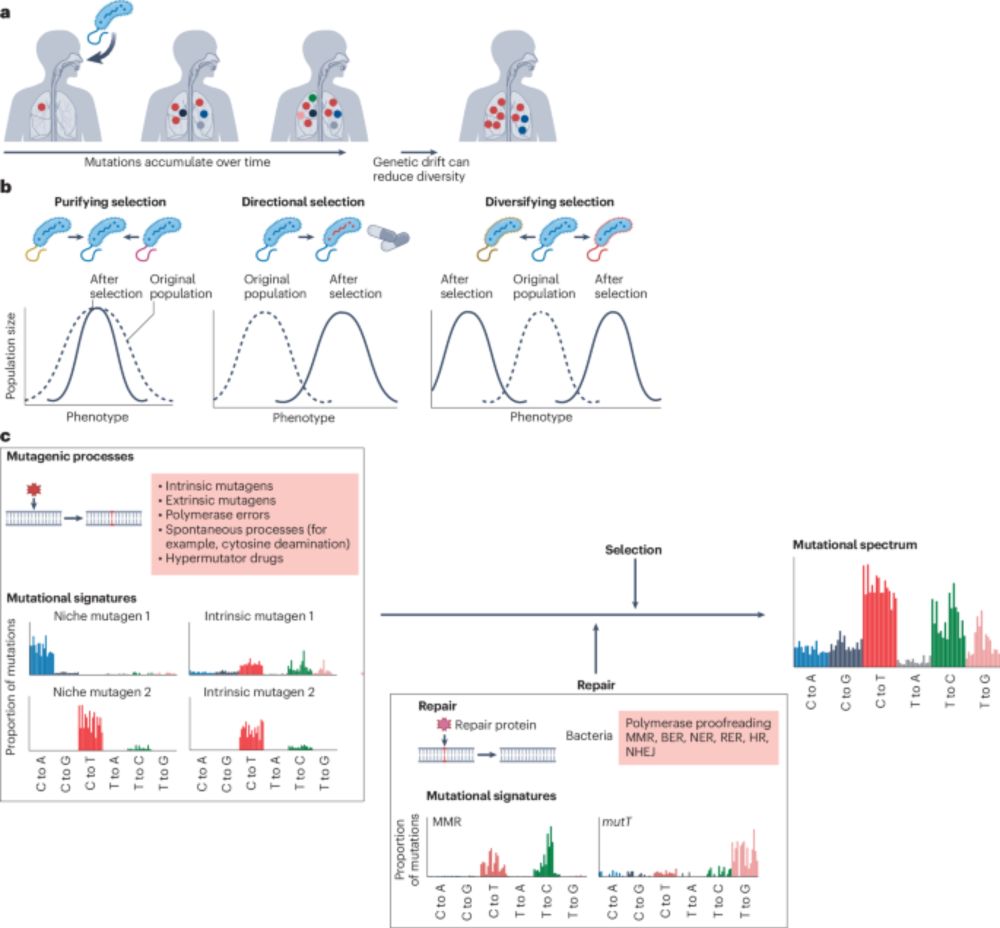
A discussion on the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host genetic diversity, and the implications for transmission analysis and infectious disease control.
#MicroSky 🦠
www.nature.com/articles/s41...

