Microbial evolution | Human microbiome ecology | Infectious disease epidemiology
We're looking for someone keen on bioinformatics and microbiome evolution.
Important info below on eligibility & URSA competition funding👇
www.findaphd.com/phds/project...

We're looking for someone keen on bioinformatics and microbiome evolution.
Important info below on eligibility & URSA competition funding👇
www.findaphd.com/phds/project...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
🧵 (1/14)
www.doi.org/10.1038/s414...
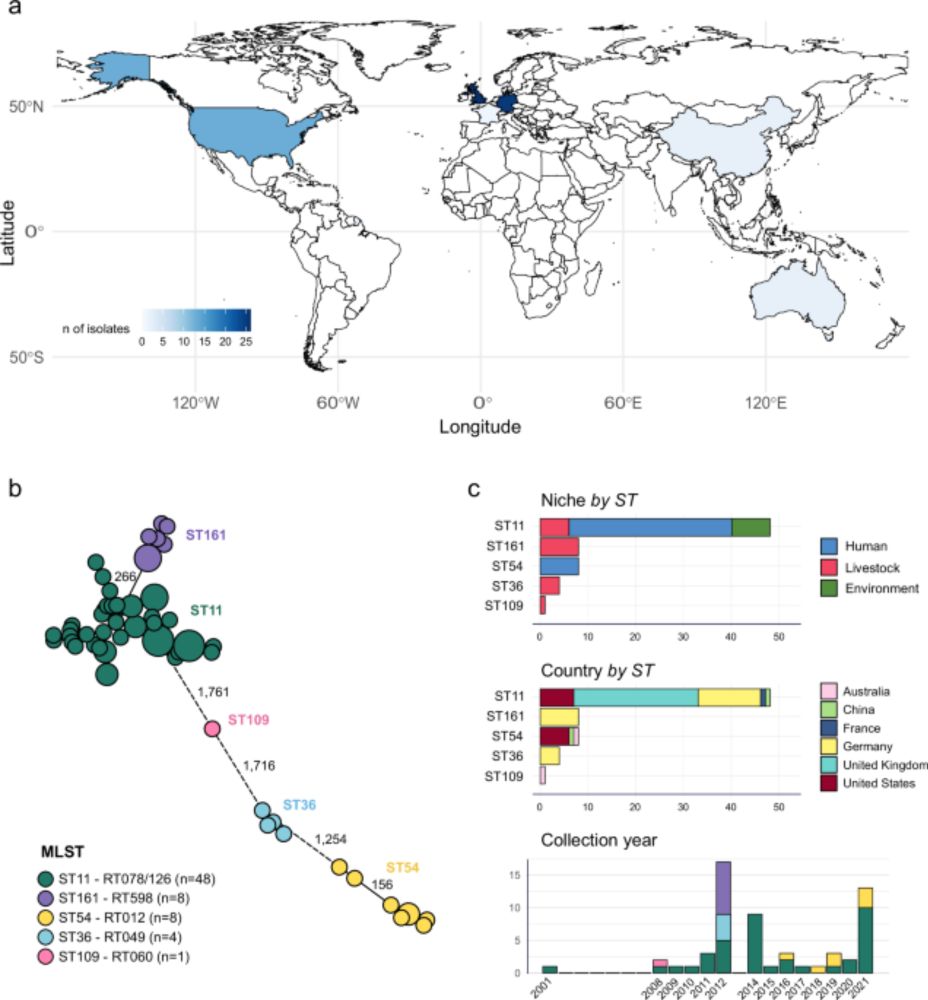
🧵 (1/14)
www.doi.org/10.1038/s414...

We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63

www.nature.com/articles/s41...

www.nature.com/articles/s41...
Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

Phages may drive microbial diversity, yet we often don’t even know how phages & bacteria correlate in nature. Our new study tackles this in the honeybee gut, thanks to the great work of PhD student @malickndiaye.bsky.social at @dmf-unil.bsky.social @fbm-unil.bsky.social

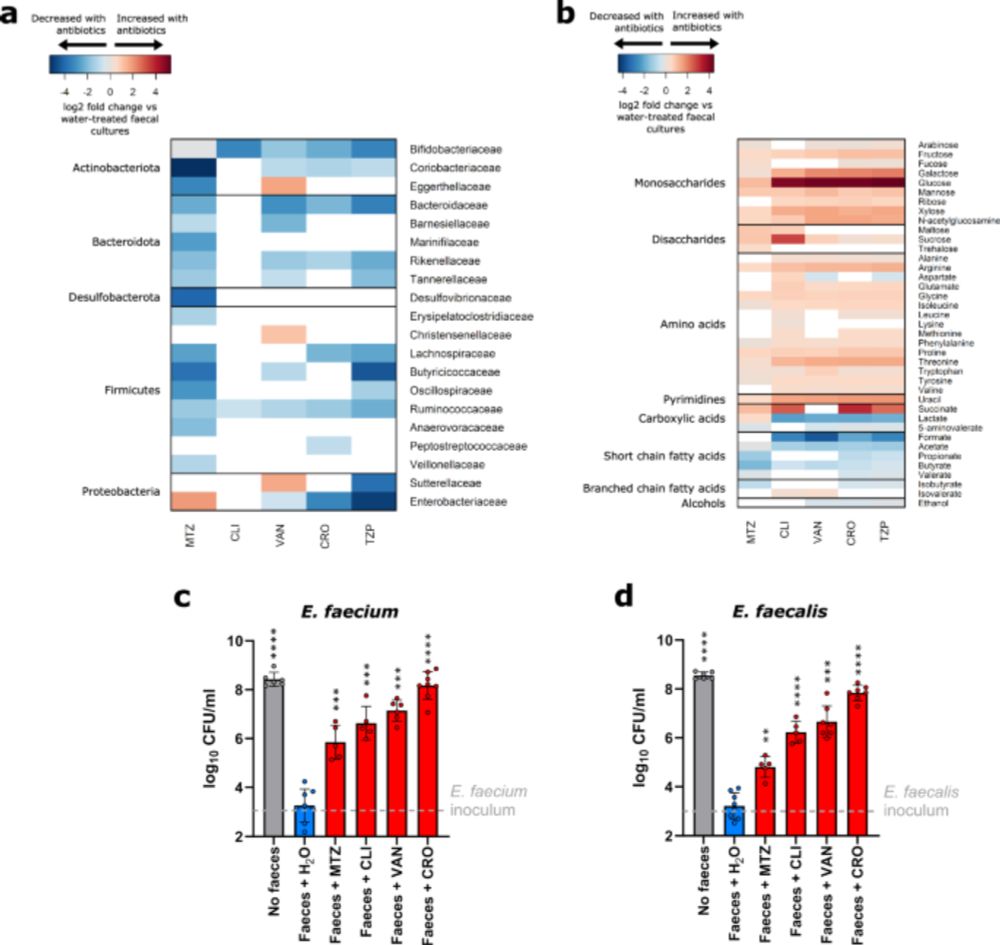


PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
PHLAME works on tough sample types -- including those with coexisting strains of a species and low depth.
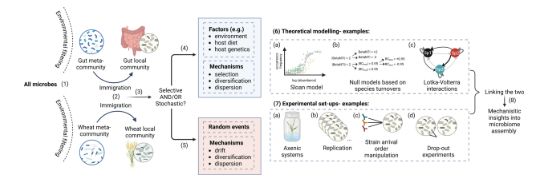
A discussion on the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host genetic diversity, and the implications for transmission analysis and infectious disease control.
#MicroSky 🦠
www.nature.com/articles/s41...
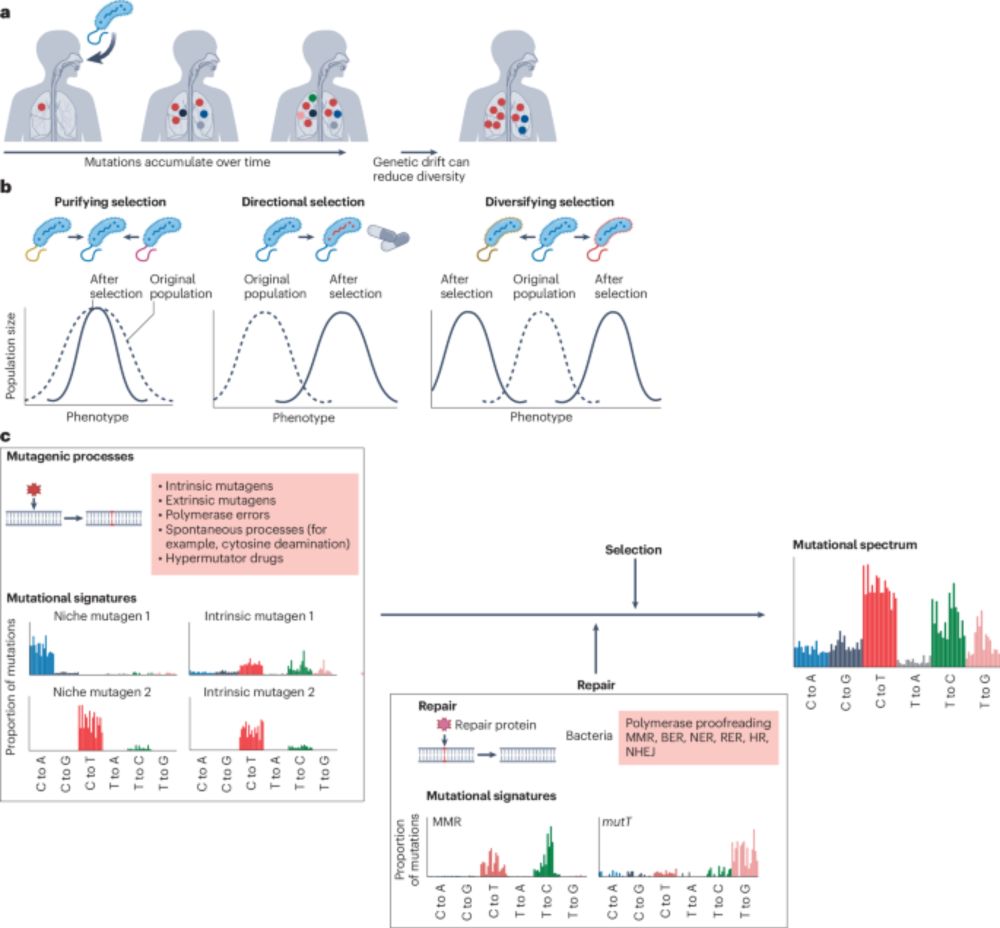
A discussion on the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host genetic diversity, and the implications for transmission analysis and infectious disease control.
#MicroSky 🦠
www.nature.com/articles/s41...
Also, please appreciate the mountain goat in Fig 1 ⛰️🐐 and that it also represents my joy for trail running and the mountains 😁
Review by Erik Bakkeren, Vit Piskovsky, and Kevin R. Foster
www.cell.com/cell-host-mi...
We show there is no close relative of Cutibacterium on the faces of gorillas and chimps at the Lincoln Park Zoo, furthering the mysterious origin of the dominant human skin colonizer.
journals.asm.org/doi/10.1128/...
We show there is no close relative of Cutibacterium on the faces of gorillas and chimps at the Lincoln Park Zoo, furthering the mysterious origin of the dominant human skin colonizer.
journals.asm.org/doi/10.1128/...

Curious about how strain diversity relates to human traits? Follow this thread! 🌍 (1/n)
www.sciencedirect.com/science/arti...
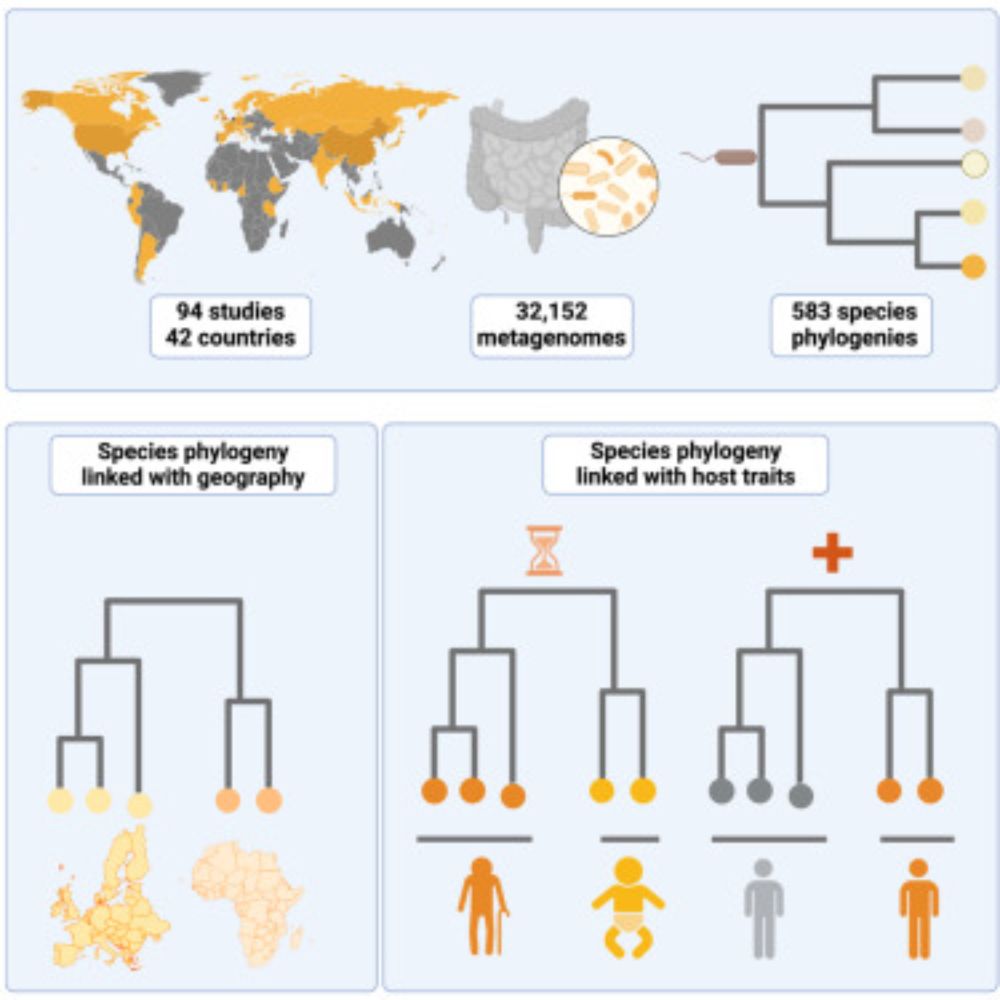
Curious about how strain diversity relates to human traits? Follow this thread! 🌍 (1/n)
www.sciencedirect.com/science/arti...
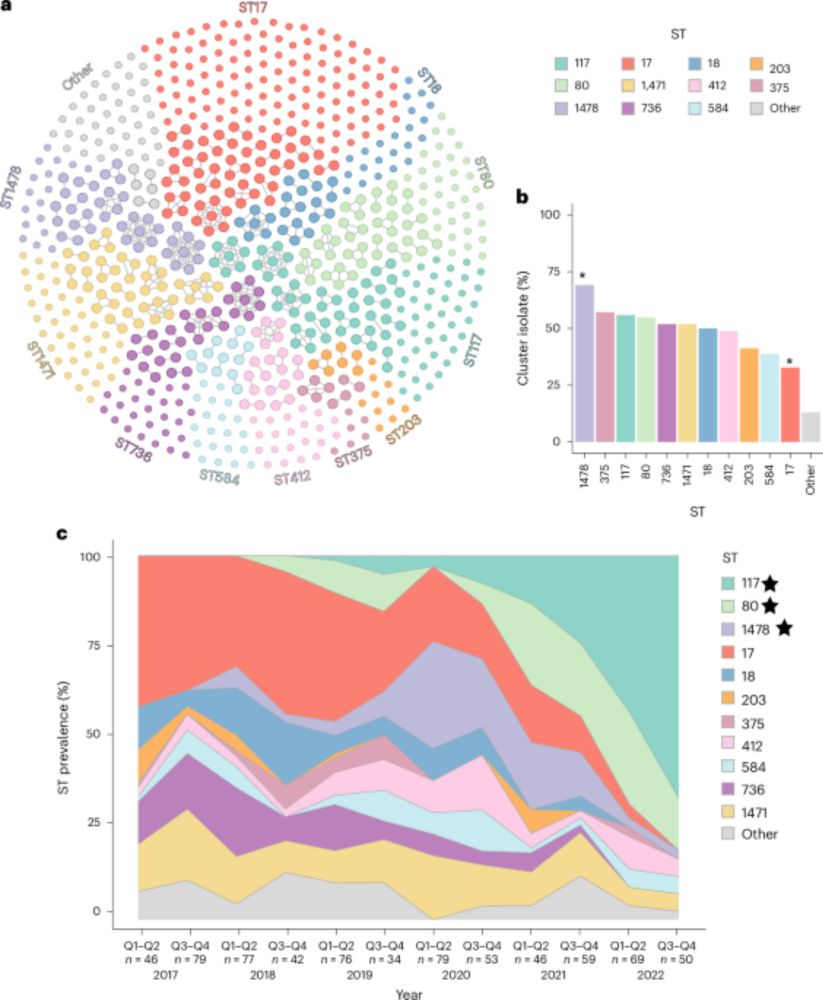
#ISMEComms by @mcastd.bsky.social, @padpadpadpad.bsky.social and Angus Buckling
academic.oup.com/ismecommun/a...

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)
"Residence time structures microbial communities through niche partitioning"
Now out in Ecology Letters:
onlinelibrary.wiley.com/doi/10.1111/...
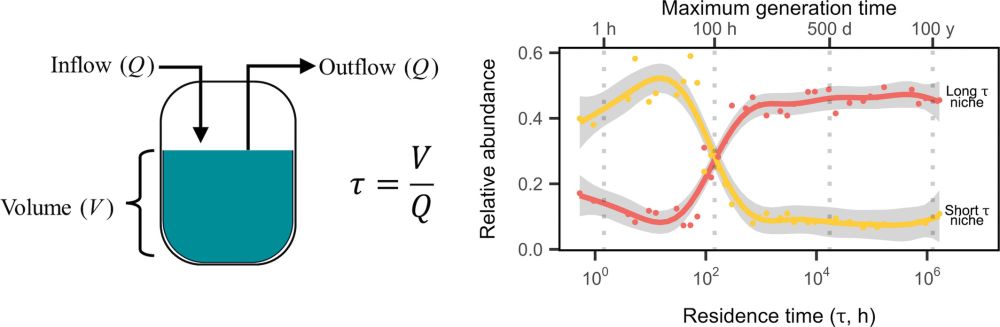
"Residence time structures microbial communities through niche partitioning"
Now out in Ecology Letters:
onlinelibrary.wiley.com/doi/10.1111/...
www.biorxiv.org/content/10.1...
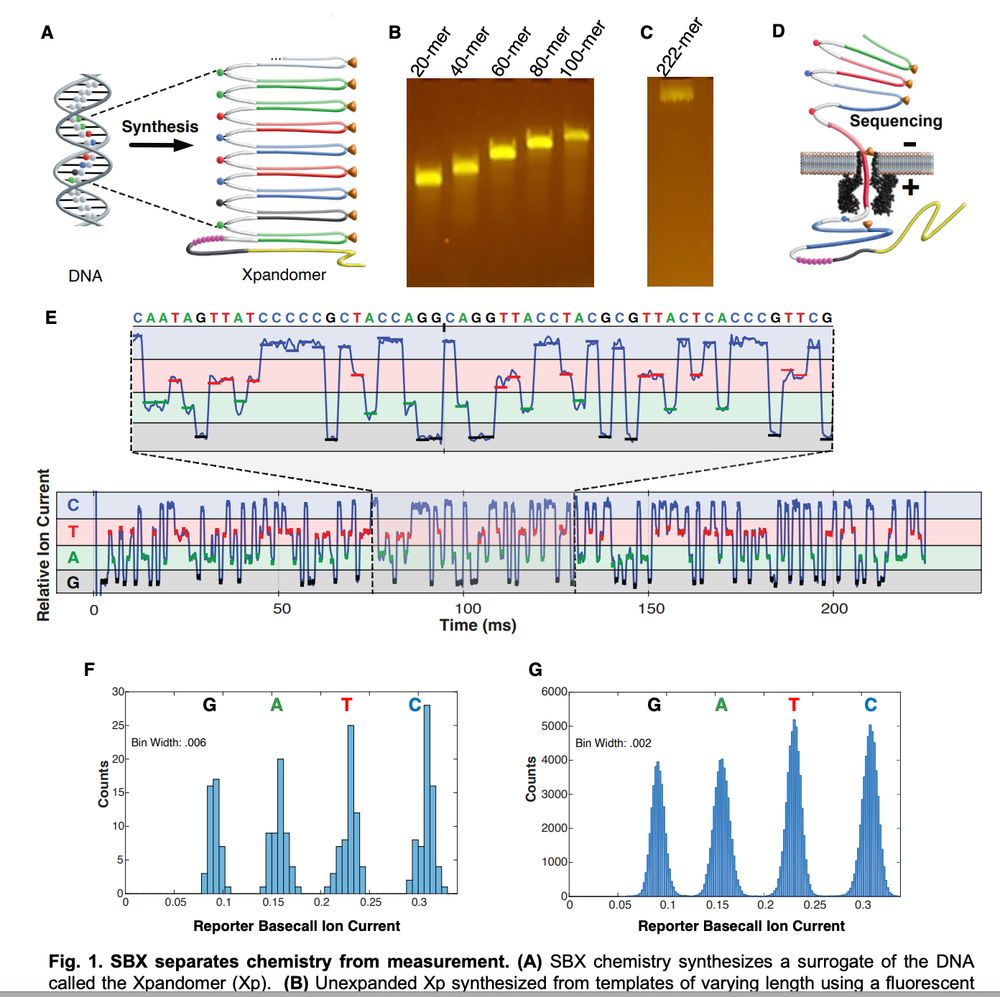
www.biorxiv.org/content/10.1...
www.fosterlab.uk/vacancies
www.fosterlab.uk/vacancies

