
| Postdoctoral researcher at IPLA-CSIC.
| Microbiology & Bioinformatics.
| RicardoLeonSampedro.com

Secret Invasion: Strain Fate Across Microbiomes
Cover illustration by Helena Klein (@illuzation.bsky.social).
communities.springernature.com/posts/secret...
Microbes construct their own niche which in turn reshapes their evolution.
Preprint drop from grad student @noahhoupt.bsky.social whose evolution experiments featured blue/white colonies, 1000 generations, a lab move, and much more!
Check out our latest preprint, now up on BioRxiv!
biorxiv.org/content/10.6...
For a quick summary, peep the thread below...🧵 (1/10)
Microbes construct their own niche which in turn reshapes their evolution.
Preprint drop from grad student @noahhoupt.bsky.social whose evolution experiments featured blue/white colonies, 1000 generations, a lab move, and much more!
Secret Invasion: Strain Fate Across Microbiomes
Cover illustration by Helena Klein (@illuzation.bsky.social).
communities.springernature.com/posts/secret...

Secret Invasion: Strain Fate Across Microbiomes
Cover illustration by Helena Klein (@illuzation.bsky.social).
communities.springernature.com/posts/secret...


Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...

Do more plasmid copies mean faster evolution?
🧵 Dive into the story
www.pnas.org/doi/10.1073/...
We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...

We represent plasmids as circles and mutations as dots, resembling an eye, because in this paper we literally 𝑤𝑎𝑡𝑐ℎ plasmids evolve.
‼️Check Paula’s 🧵 and the paper👇
𝗣𝗹𝗮𝘀𝗺𝗶𝗱 𝗺𝘂𝘁𝗮𝘁𝗶𝗼𝗻 𝗿𝗮𝘁𝗲𝘀 𝘀𝗰𝗮𝗹𝗲 𝘄𝗶𝘁𝗵 𝗰𝗼𝗽𝘆 𝗻𝘂𝗺𝗯𝗲𝗿
www.pnas.org/doi/10.1073/...
Looking to recruit a postdoc to join a UKRI FLF-funded project on antibiotic resistance evolution in the microbiome 🦠
3 years funding, deadline 26th Jan, please share!
hrwebapp.qub.ac.uk/tlive_webrec...


We found that loss-of-function mutations in the carbapenem entry porin OprD of Pseudomonas aeruginosa do more than confer #AntibioticResistance: they reshape the bacterial membrane and interaction with the host, enhancing epithelial colonization capacity 🦠
#Microsky
We found that loss-of-function mutations in the carbapenem entry porin OprD of Pseudomonas aeruginosa do more than confer #AntibioticResistance: they reshape the bacterial membrane and interaction with the host, enhancing epithelial colonization capacity 🦠
#Microsky

#MicroSky #Mevosky
journals.plos.org/plosbiology/...

#MicroSky #Mevosky
journals.plos.org/plosbiology/...
(1/n) www.biorxiv.org/content/earl...

(1/n) www.biorxiv.org/content/earl...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

With #HealthResearch support, @sanmillan.bsky.social is investigating how different plasmids (DNA fragments) coexist in bacteria and confer resistance.
https://tinyurl.com/3bysv6cr

With #HealthResearch support, @sanmillan.bsky.social is investigating how different plasmids (DNA fragments) coexist in bacteria and confer resistance.
https://tinyurl.com/3bysv6cr
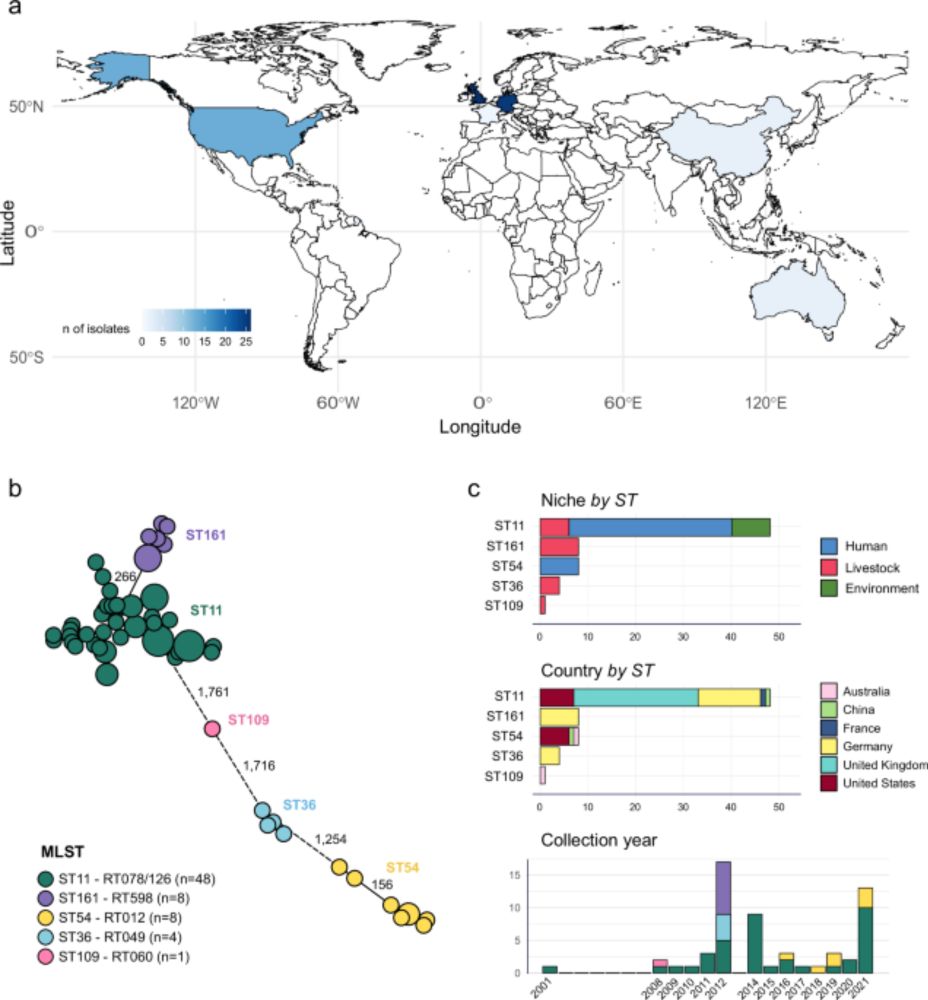
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63


🧵 (1/14)
www.doi.org/10.1038/s414...
🧵 (1/14)
www.doi.org/10.1038/s414...
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63

We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63

Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...

Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...

Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
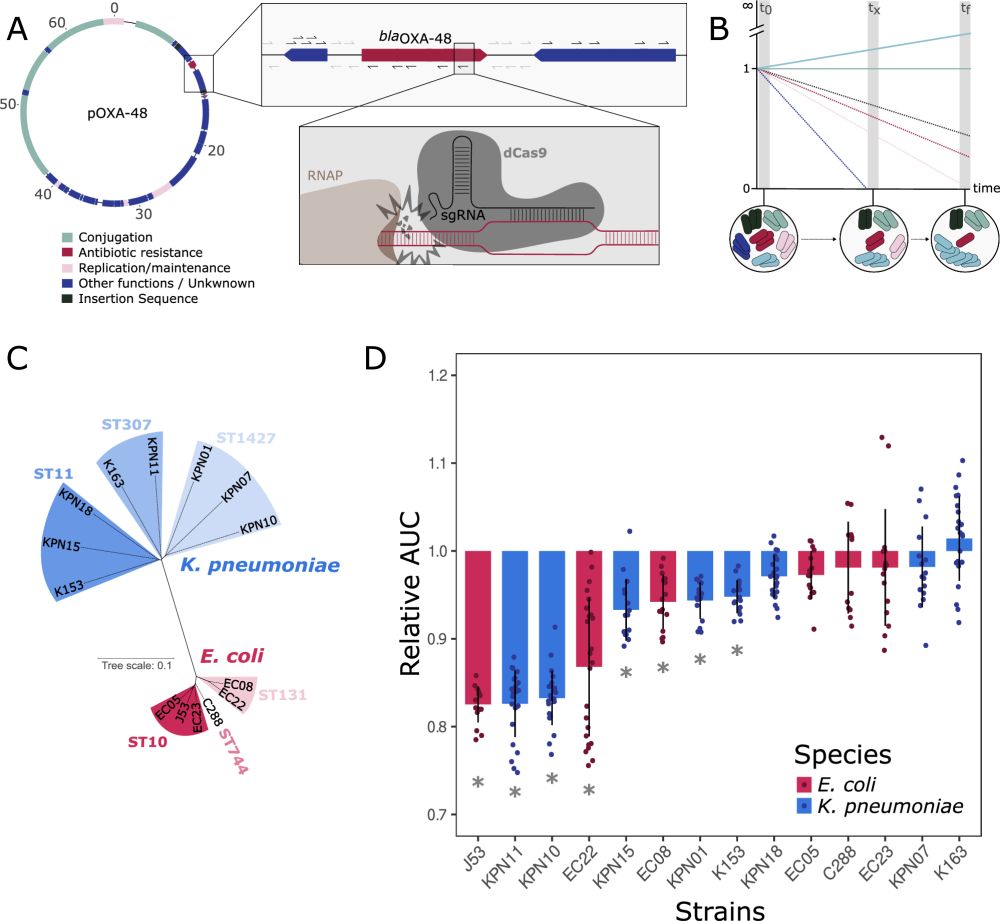
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
www.nature.com/articles/s41...

www.nature.com/articles/s41...

