In this preprint, Lucas Paoli et al, ask what shapes antiphage systems expression in native contexts.
www.biorxiv.org/content/10.6...

In this preprint, Lucas Paoli et al, ask what shapes antiphage systems expression in native contexts.
www.biorxiv.org/content/10.6...

A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
www.pnas.org/doi/10.1073/...

www.pnas.org/doi/10.1073/...
🧫🦠
In oysters, some stay identical for years.
With >1,200 phages & 600 Vibrio genomes, we reveal long-term stability and new mobile elements.
Proud of this collaborative work across our teams (Roscoff-UdeM and @epcrocha.bsky.social www.biorxiv.org/cgi/content/...

🧫🦠
www.nature.com/articles/s41...

www.nature.com/articles/s41...
That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇

That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇
🧬 He's pioneering phage-CRISPR tools to edit gut #bacteria in vivo, reshaping the #microbiome from within and opening the door to targeted therapies.
👉 europa.eu/!HQbHTw
👉 europa.eu/!KtWf33
@dbikard.bsky.social



We analyze KP-phage interactions from an eco-evo perspective, summarize many lab and clinical trials, and discuss novel approaches like genetic engineering & machine learning.
#microsky
doi.org/10.1371/jour...
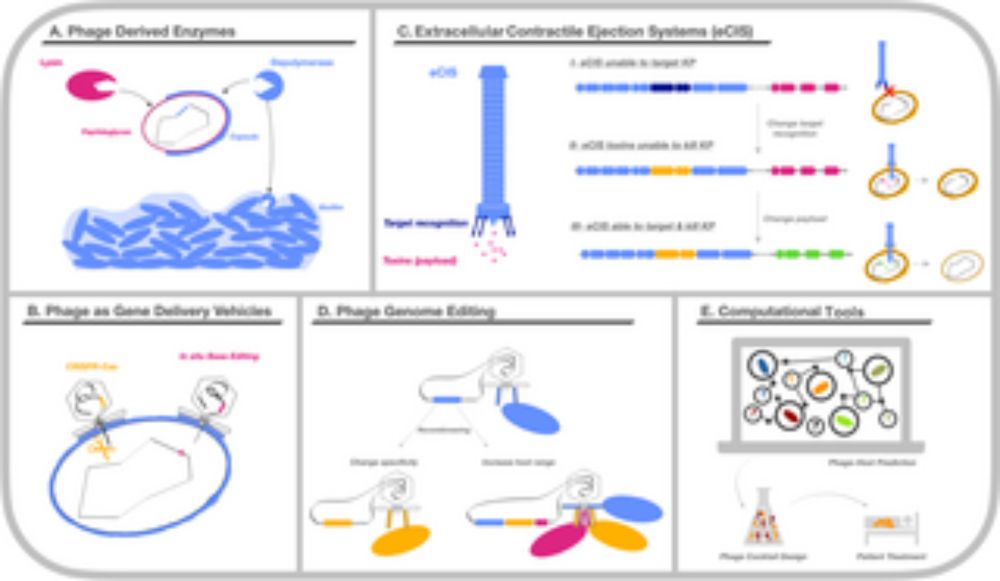
We analyze KP-phage interactions from an eco-evo perspective, summarize many lab and clinical trials, and discuss novel approaches like genetic engineering & machine learning.
#microsky
doi.org/10.1371/jour...



