
Cool to see people trying it out, find it on @bioconductor.bsky.social if you want to give it a go bioconductor.org/packages//re...
Cool to see people trying it out, find it on @bioconductor.bsky.social if you want to give it a go bioconductor.org/packages//re...
The NOMIS Synergy Fellowship offers a great opportunity to develop such a position in my lab within the interdisciplinary Synergy excellence cluster.
🔗 www.synergy-munich.de/news-events/...

The NOMIS Synergy Fellowship offers a great opportunity to develop such a position in my lab within the interdisciplinary Synergy excellence cluster.
🔗 www.synergy-munich.de/news-events/...

www.nature.com/articles/s41...

www.nature.com/articles/s41...


Help us build large-scale bio-inspired neural networks, write high-quality research code, and contribute to open-source tools like jaxley, sbi, and flyvis 🪰.
More info: www.mackelab.org/jobs/
Help us build large-scale bio-inspired neural networks, write high-quality research code, and contribute to open-source tools like jaxley, sbi, and flyvis 🪰.
More info: www.mackelab.org/jobs/




Erika Alden DeBenedictis, Co-founder of The Align Foundation 🧵
@erika-alden.bsky.social
@alignbio.bsky.social
@pioneerlabs.bsky.social
#scverse #scverse2025 #StructuralBiology #Biotech #Stanford

Erika Alden DeBenedictis, Co-founder of The Align Foundation 🧵
@erika-alden.bsky.social
@alignbio.bsky.social
@pioneerlabs.bsky.social
#scverse #scverse2025 #StructuralBiology #Biotech #Stanford
We're delighted to recognize 10x Genomics as a Platinum Sponsor of scverse conference 2025! 🏆
@10xgenomics.bsky.social
#scverse2025 #ComputationalBiology #SingleCell #SpatialOmics #10xGenomics #Bioinformatics #Biotechnology


We're delighted to recognize 10x Genomics as a Platinum Sponsor of scverse conference 2025! 🏆
@10xgenomics.bsky.social
#scverse2025 #ComputationalBiology #SingleCell #SpatialOmics #10xGenomics #Bioinformatics #Biotechnology
If you're a PI working in algorithmic genomics (& you can recommend my lab to your top graduating students ;P), please let them know!
Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social

Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social

academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
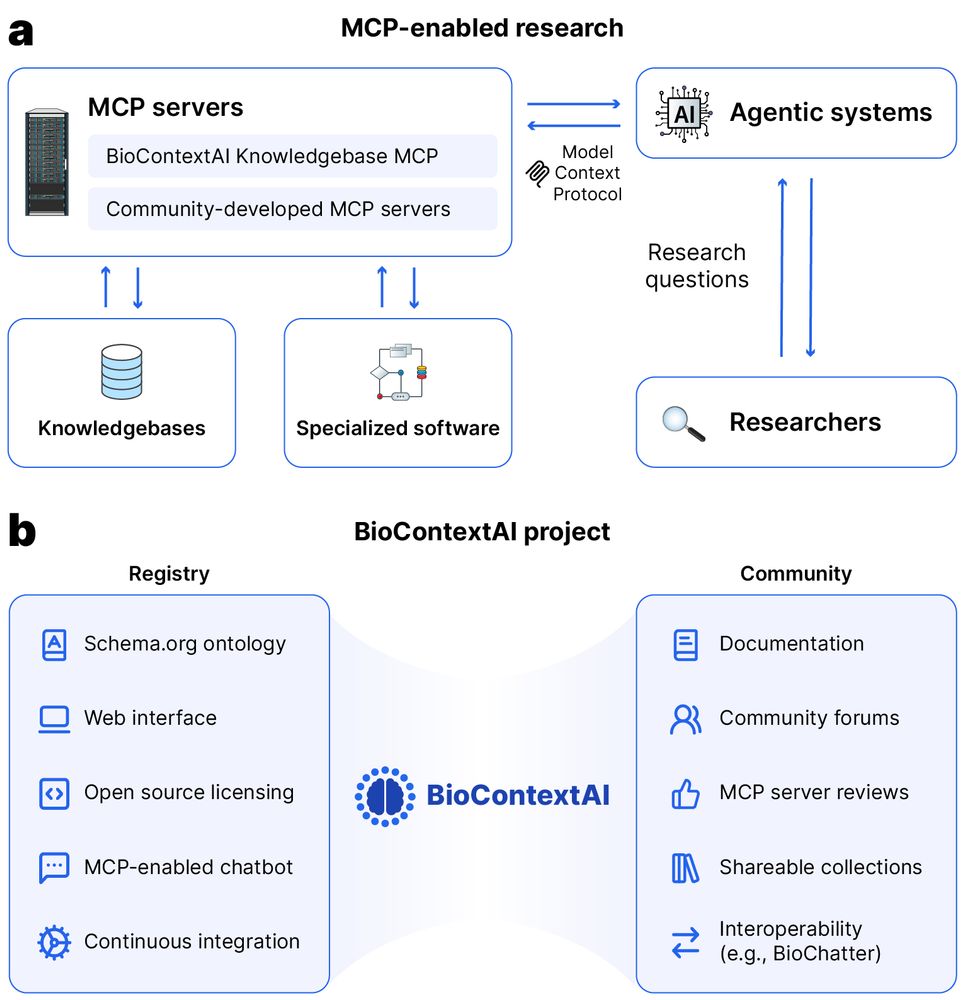
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
(Zoom registration link and more information in thread!)

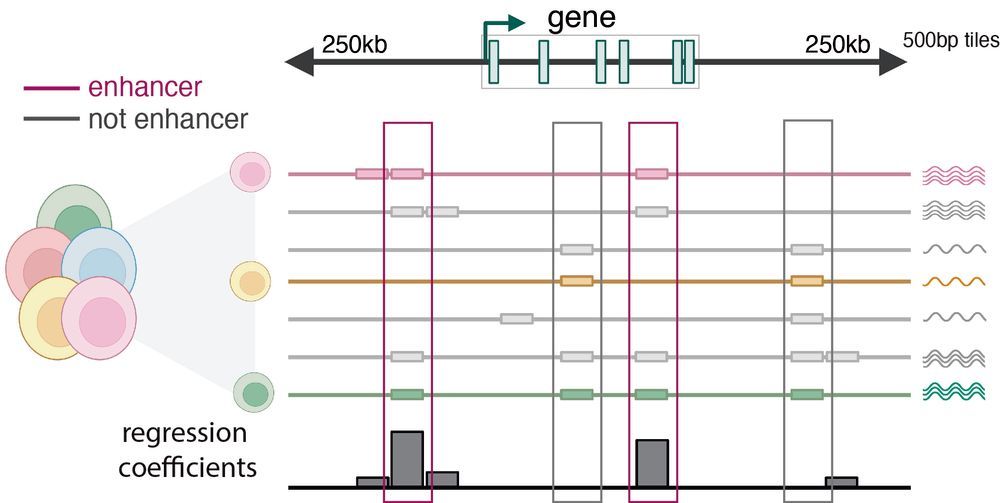
(Zoom registration link and more information in thread!)
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
Inspired by ideas in www.nature.com/articles/s41... and github.com/VPetukhov/GP..., implemented as an scVerse ecosystem package with docs and tutorials.
Inspired by ideas in www.nature.com/articles/s41... and github.com/VPetukhov/GP..., implemented as an scVerse ecosystem package with docs and tutorials.
Anastasia Litinetskaya will present “Multimodal integration and identification of disease-specific changes in single-cell atlases”. Check out the GitHub repo here: github.com/theislab/mul...
Anastasia Litinetskaya will present “Multimodal integration and identification of disease-specific changes in single-cell atlases”. Check out the GitHub repo here: github.com/theislab/mul...
Anastasia Litinetskaya will present “Multimodal integration and identification of disease-specific changes in single-cell atlases”. Check out the GitHub repo here: github.com/theislab/mul...

Free to read here: rdcu.be/eabUB
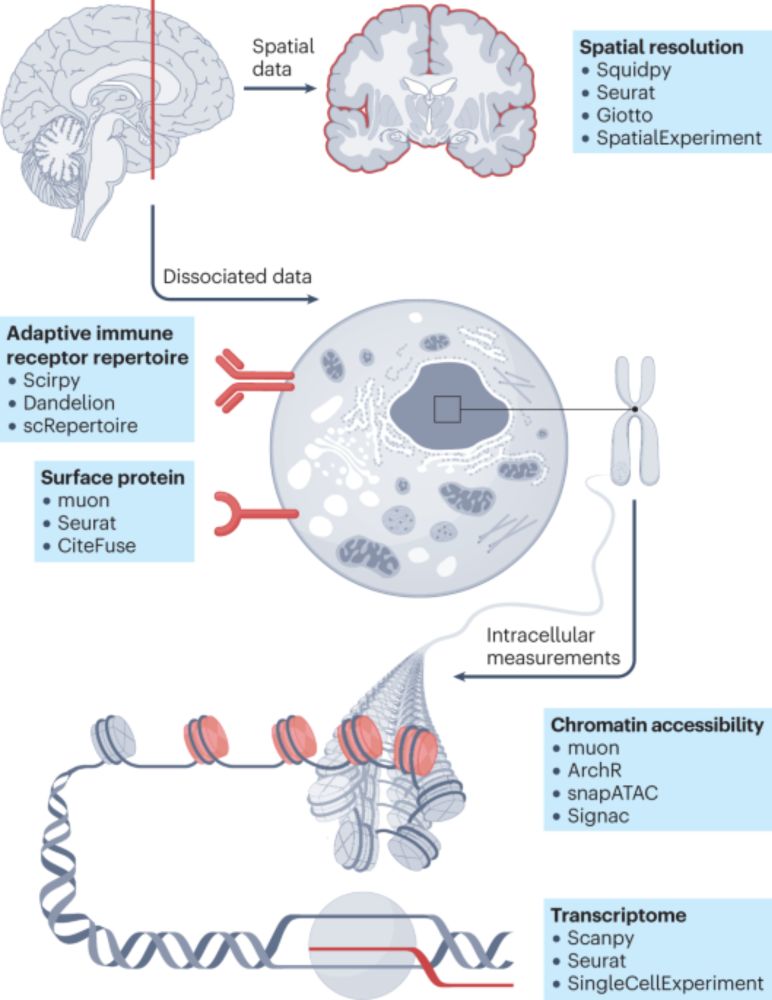
Free to read here: rdcu.be/eabUB


