
☕️🎧🇦🇺🎥

Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...

Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...
bsky.app/profile/rauf...
Github: github.com/Kalan-Lab/zol
Docs: github.com/Kalan-Lab/zo...
#secmet #phage #genomics
bsky.app/profile/rauf...
@nfrk92.bsky.social
@narjournal.bsky.social
academic.oup.com/nar/article/...

@nfrk92.bsky.social
@narjournal.bsky.social
academic.oup.com/nar/article/...
doi.org/10.1098/rstb...

doi.org/10.1098/rstb...
#microsky #microbiomesky

#microsky #microbiomesky

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
"Plasmids, prophages, and defense systems are depleted from plant microbiota genomes"
genomebiology.biomedcentral.com/articles/10....
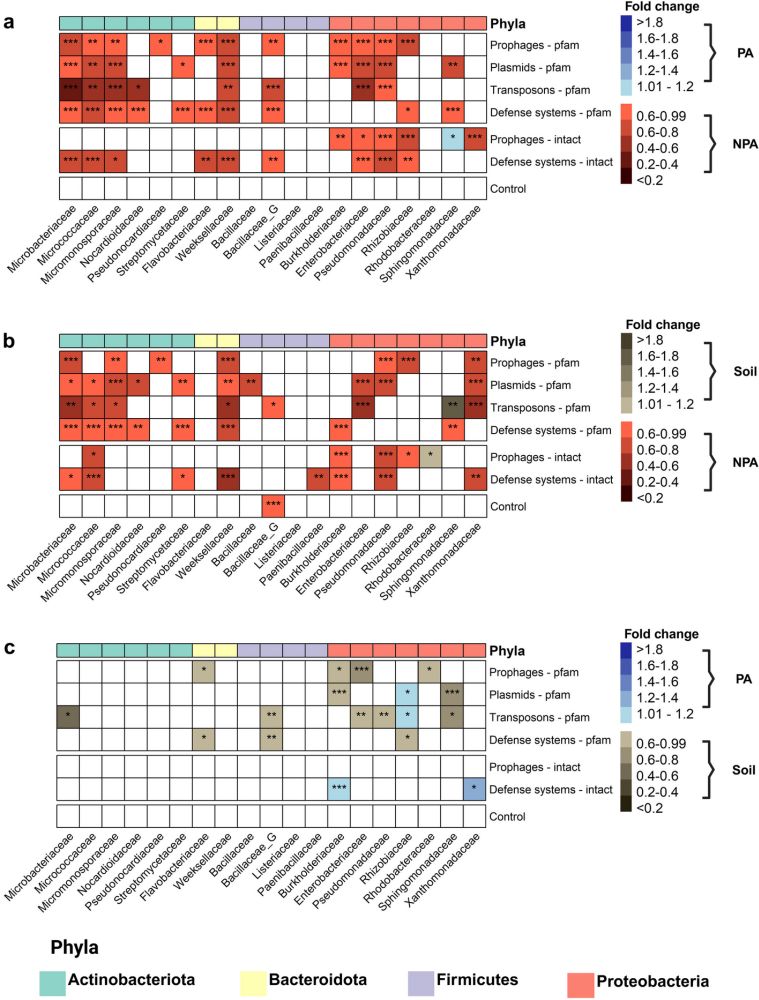
"Plasmids, prophages, and defense systems are depleted from plant microbiota genomes"
genomebiology.biomedcentral.com/articles/10....
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
Autocycler, the automated successor to Trycycler from @rrwick.bsky.social has a pre-print out - overall, pretty awesome performance (and is very easy to use)
github.com/rrwick/Autoc...

Autocycler, the automated successor to Trycycler from @rrwick.bsky.social has a pre-print out - overall, pretty awesome performance (and is very easy to use)
github.com/rrwick/Autoc...
We describe a novel, cultivated #Archaea species from the human microbiome: #Methanobrevibacter intestini sp. nov. !
www.microbiologyresearch.org/content/jour...
#Microbiome #GutMicrobiota #NewSpecies #archaeasky #microbiomesky

We describe a novel, cultivated #Archaea species from the human microbiome: #Methanobrevibacter intestini sp. nov. !
www.microbiologyresearch.org/content/jour...
#Microbiome #GutMicrobiota #NewSpecies #archaeasky #microbiomesky
#microsky 🧪
www.nature.com/articles/s41...
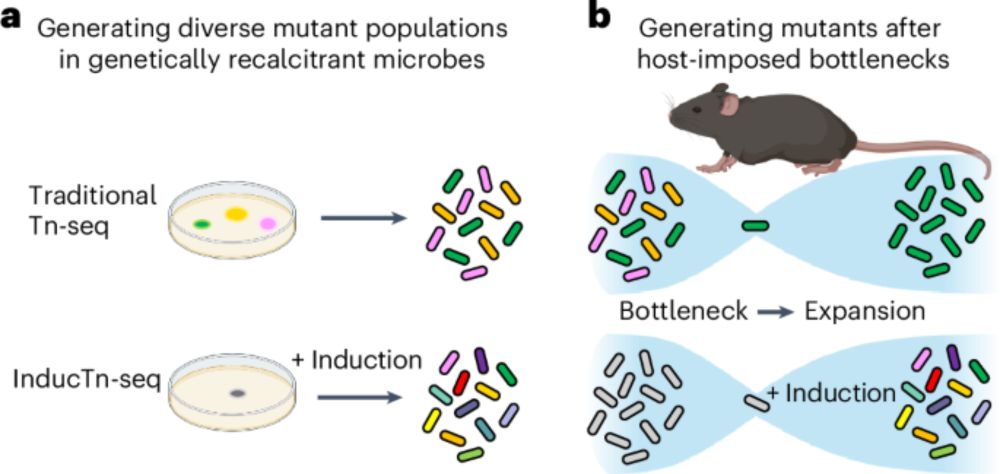
#microsky 🧪
www.nature.com/articles/s41...
Also suitable to build saturated chromosomal variant libraries using oligo-recombineering with extra short homology arms!
www.biorxiv.org/content/10.1...

Also suitable to build saturated chromosomal variant libraries using oligo-recombineering with extra short homology arms!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Start your career in ground-breaking research with Gal Ofir & Christian Rödelsperger from the MPI Biology Tübingen.
Apply by: 27 Jan. www.phd.tuebingen.mp...
#imprs #graduateprogram #maxplanck #phd #phdlife #phdposition #phdpositiongermany #biology


Start your career in ground-breaking research with Gal Ofir & Christian Rödelsperger from the MPI Biology Tübingen.
Apply by: 27 Jan. www.phd.tuebingen.mp...
#imprs #graduateprogram #maxplanck #phd #phdlife #phdposition #phdpositiongermany #biology

"Extensive impact of non-antibiotic drugs on human gut bacteria" (2018) www.nature.com/artic...

"Extensive impact of non-antibiotic drugs on human gut bacteria" (2018) www.nature.com/artic...
#Microsky #phagesky #Science
ntu.wd3.myworkdayjobs.com/Careers/job/...

#Microsky #phagesky #Science
ntu.wd3.myworkdayjobs.com/Careers/job/...
🔁 Please RT & ❤️ this post so we can gather all the microbiome enthusiasts and scientists in one place. #MicrobiomeScience #Networking
🔁 Please RT & ❤️ this post so we can gather all the microbiome enthusiasts and scientists in one place. #MicrobiomeScience #Networking
If you are interested in marine microbiology, bacterial genetics and/or symbiosis, please consider reaching out! Repost to help spread the word 🪸🧪🧬🌊
If you are interested in marine microbiology, bacterial genetics and/or symbiosis, please consider reaching out! Repost to help spread the word 🪸🧪🧬🌊


www.nature.com/articles/s41...

www.nature.com/articles/s41...


