CryoET everything! #TeamTomo
We extend our Surface Morphometrics pipeline to measure membrane thickness.
Alongside many biological findings, we show that our 3D measurements closely match in vitro vesicle measurements from 2D EM (collab w/Neal Waxham and Fred Heberle).
#CryoET

We extend our Surface Morphometrics pipeline to measure membrane thickness.
Alongside many biological findings, we show that our 3D measurements closely match in vitro vesicle measurements from 2D EM (collab w/Neal Waxham and Fred Heberle).


s/o to co-authors @baradlab.com @hamid13r.bsky.social @zidlab.bsky.social
#teamtomo #cellbio25
📕 From The Year In Cell Biology: rupress.org/jcb/collecti...
#CellBio2025

s/o to co-authors @baradlab.com @hamid13r.bsky.social @zidlab.bsky.social
#teamtomo #cellbio25
I met Matt Iadanza at the CZI but I guess he doesn't have bluesky to tag!

I met Matt Iadanza at the CZI but I guess he doesn't have bluesky to tag!
Lab all-star @mmedina300kv.bsky.social maps these microdomains using cryo-ET + surface morphometrics.
s/o to #teamtomo #cryoET co-authors
@hamid13r.bsky.social
@attychang.bsky.social
@baradlab.com
Lab all-star @mmedina300kv.bsky.social maps these microdomains using cryo-ET + surface morphometrics.
s/o to #teamtomo #cryoET co-authors
@hamid13r.bsky.social
@attychang.bsky.social
@baradlab.com
Lab all-star @mmedina300kv.bsky.social maps these microdomains using cryo-ET + surface morphometrics.
s/o to #teamtomo #cryoET co-authors
@hamid13r.bsky.social
@attychang.bsky.social
@baradlab.com

mrc.tal.net/vx/appcentre...
mrc.tal.net/vx/appcentre...
rm -rf ~/
github.com/hamid13r/war...

github.com/hamid13r/war...
I wrote a python code to turn the UseTilt to False in the XML files for those views and improve the tomo reconstruction.
#teamtomo
github.com/hamid13r/war...

I wrote a python code to turn the UseTilt to False in the XML files for those views and improve the tomo reconstruction.
#teamtomo
github.com/hamid13r/war...
This looks like a fantastic meeting!
www.nexperion.net/semcm2026

This looks like a fantastic meeting!


Unfortunately, as an immigrant I would rather not to explain any further.
Unfortunately, as an immigrant I would rather not to explain any further.
Open #PhDposition in structural virology in my group! Join us in Umeå, 🇸🇪 to uncover how arboviruses remodel the cellular interior. In situ #cryoET combined with #virology, #cellbiology, and #biophysics.
Deadline 4 May. More info: www.carlsonlab.se/join/
We expanded Surface Morphometrics to quantify membrane thickness from cryo-ET—revealing local variation across organelles.
Led by the lab’s first grad student, @mmedina300kv.bsky.social (defending Monday! 🍾) w/ @attychang.bsky.social @hamid13r.bsky.social & @tomo.science
We expanded Surface Morphometrics to quantify membrane thickness from cryo-ET—revealing local variation across organelles.
Led by the lab’s first grad student, @mmedina300kv.bsky.social (defending Monday! 🍾) w/ @attychang.bsky.social @hamid13r.bsky.social & @tomo.science
The cover shows a segmented model of cytoplasmic #ribosomes associated with mitochondrial membrane in a Saccharomyces cerevisiae cell. From Ya-Ting Chang, @nanigrotjahn.bsky.social and colleagues (doi.org/10.1083/jcb....)

Open #PhDposition in structural virology in my group! Join us in Umeå, 🇸🇪 to uncover how arboviruses remodel the cellular interior. In situ #cryoET combined with #virology, #cellbiology, and #biophysics.
Deadline 4 May. More info: www.carlsonlab.se/join/
Open #PhDposition in structural virology in my group! Join us in Umeå, 🇸🇪 to uncover how arboviruses remodel the cellular interior. In situ #cryoET combined with #virology, #cellbiology, and #biophysics.
Deadline 4 May. More info: www.carlsonlab.se/join/
Suddenly, I remembered Atty's paper:
Not only we can locate the ribosomes, we can see their orientations, on the surface of the mitochondria outer membrane!
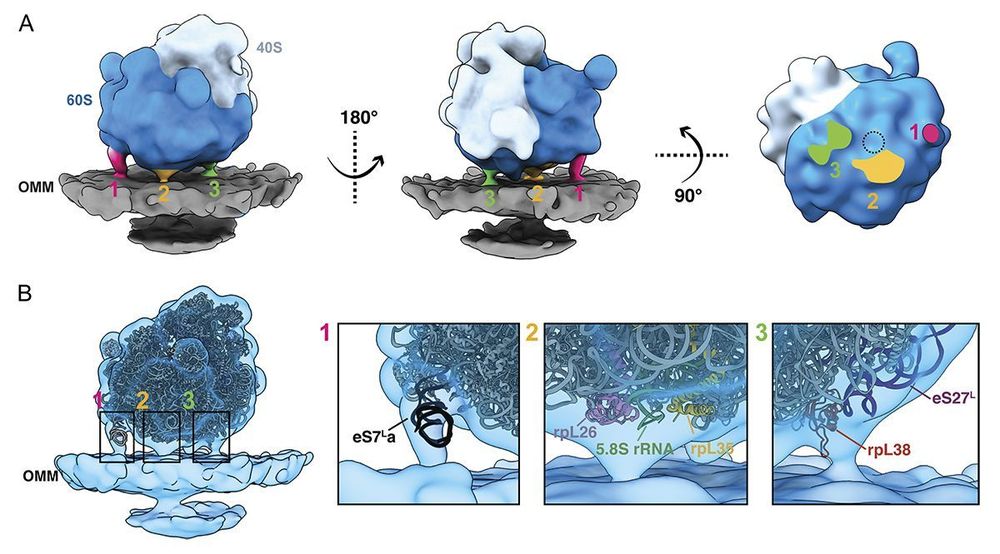
Suddenly, I remembered Atty's paper:
Not only we can locate the ribosomes, we can see their orientations, on the surface of the mitochondria outer membrane!


