PhD Coyle lab https://www.coylelab.org/
Postdoc Rosen lab
A new coarse-grained model that probes full chromatin condensates at near-atomistic resolution to reveal the molecular regulation of chromatin structure and phase separation
Brilliantly led by @kieran-russell.bsky.social, with the Rosen and Orozco groups

A new coarse-grained model that probes full chromatin condensates at near-atomistic resolution to reveal the molecular regulation of chromatin structure and phase separation
Brilliantly led by @kieran-russell.bsky.social, with the Rosen and Orozco groups
www.science.org/doi/10.1126/...
@unmc.bsky.social @uwbiochem.bsky.social


Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
In the past, we have learnt that Oct4 can induce nucleosome breathing on the mono-nucleosome level.
But what happens when you have a fibre of multiple nucleosomes?
www.biorxiv.org/content/10.1...
@rcollepardo.bsky.social @juliamaristany.bsky.social


In the past, we have learnt that Oct4 can induce nucleosome breathing on the mono-nucleosome level.
But what happens when you have a fibre of multiple nucleosomes?
www.biorxiv.org/content/10.1...
@rcollepardo.bsky.social @juliamaristany.bsky.social

In the past, we have learnt that Oct4 can induce nucleosome breathing on the mono-nucleosome level.
But what happens when you have a fibre of multiple nucleosomes?
www.biorxiv.org/content/10.1...
@rcollepardo.bsky.social @juliamaristany.bsky.social
In collaboration between the Rosen, Redding, Collepardo-Guevara & Gerlich labs, we uncover a surprising principle of chromosome organisation: electrostatic repulsion positions centromeres at the chromosome surface during mitosis.
🔗 doi.org/10.1101/2025...

In collaboration between the Rosen, Redding, Collepardo-Guevara & Gerlich labs, we uncover a surprising principle of chromosome organisation: electrostatic repulsion positions centromeres at the chromosome surface during mitosis.
🔗 doi.org/10.1101/2025...

www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)

www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...


New reactions are typically developed by trial and error. How can we speed up this process? Read on to learn how we used DNA scaffolding to perform >500,000 parallel reactions on attomole scale.
1/n
New reactions are typically developed by trial and error. How can we speed up this process? Read on to learn how we used DNA scaffolding to perform >500,000 parallel reactions on attomole scale.
1/n

www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
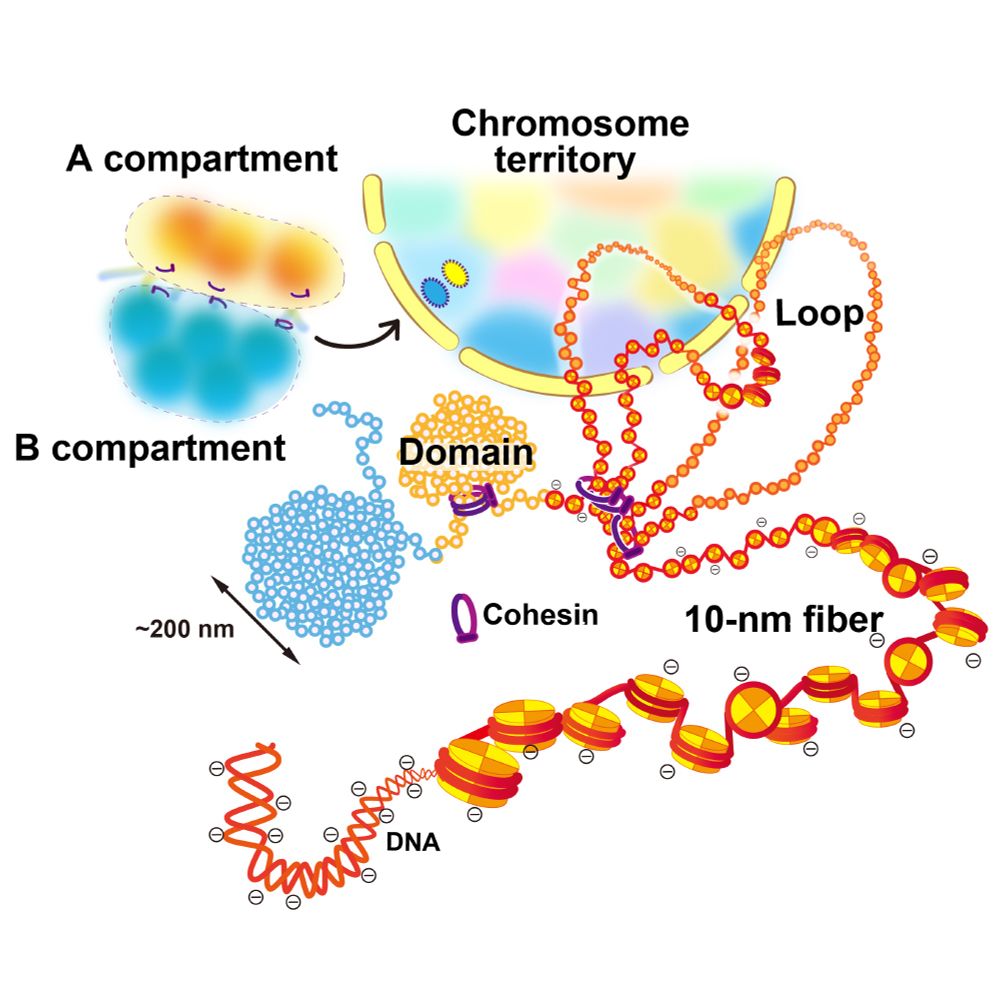
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
We decided to call them TIGR-Tas systems 🐯
We decided to call them TIGR-Tas systems 🐯
We introduce TIGR-Tas, a PAM-free, compact, modular RNA-guided DNA-targeting system in prokaryotes & their viruses.
More details coming soon 👀
#TIGR #RNA #GeneEditing #CRISPR
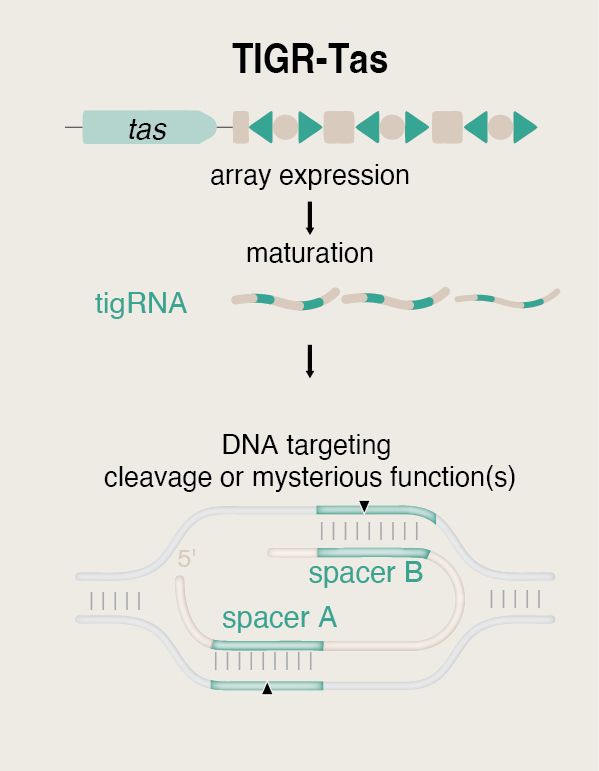
We introduce TIGR-Tas, a PAM-free, compact, modular RNA-guided DNA-targeting system in prokaryotes & their viruses.
More details coming soon 👀
#TIGR #RNA #GeneEditing #CRISPR