www.matrixscience.com/blog/top-3-q...
#proteomics #massspec
www.matrixscience.com/blog/top-3-q...
#proteomics #massspec
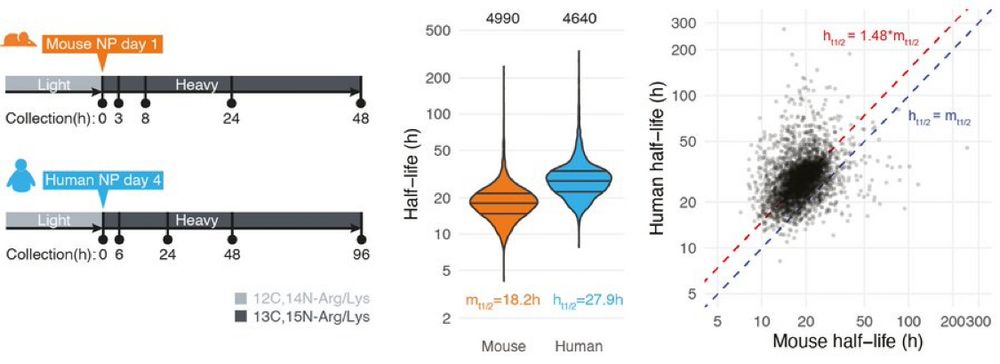

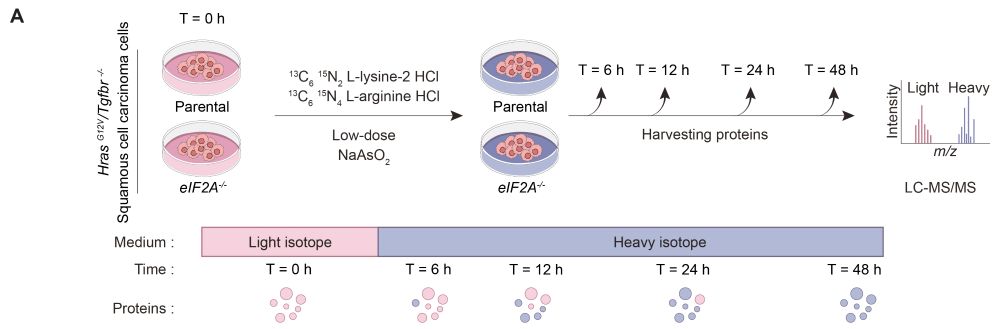
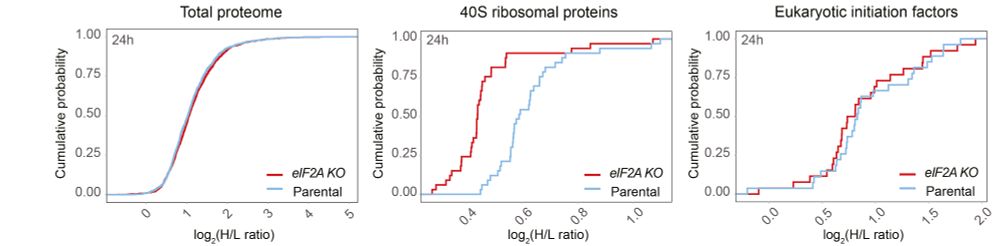
Learn more: bit.ly/3VIWkpd

Learn more: bit.ly/3VIWkpd
In a groundbreaking proteogenomic study, researchers have unveiled the profound impact of missense mutations on protein abundance within prostate cancer tissues, employing a sophisticated Stable…

In a groundbreaking proteogenomic study, researchers have unveiled the profound impact of missense mutations on protein abundance within prostate cancer tissues, employing a sophisticated Stable…
An algorithm for peptide de novo sequencing from a group of SILAC labeled MS/MS spectra - PubMed pubmed.ncbi.nlm.nih....
---
#proteomics #prot-paper

An algorithm for peptide de novo sequencing from a group of SILAC labeled MS/MS spectra - PubMed pubmed.ncbi.nlm.nih....
---
#proteomics #prot-paper
Liberty Mutual
Spectrum
Quicken
Silac
Shopify
Sterns and Foster
American Home Shield
Pure Chill
Sling
CITI
SimpliSafe
Progressive
ADT
Denver Mattress
Select Quote
Invisible Glass
Liberty Mutual
Spectrum
Quicken
Silac
Shopify
Sterns and Foster
American Home Shield
Pure Chill
Sling
CITI
SimpliSafe
Progressive
ADT
Denver Mattress
Select Quote
Invisible Glass
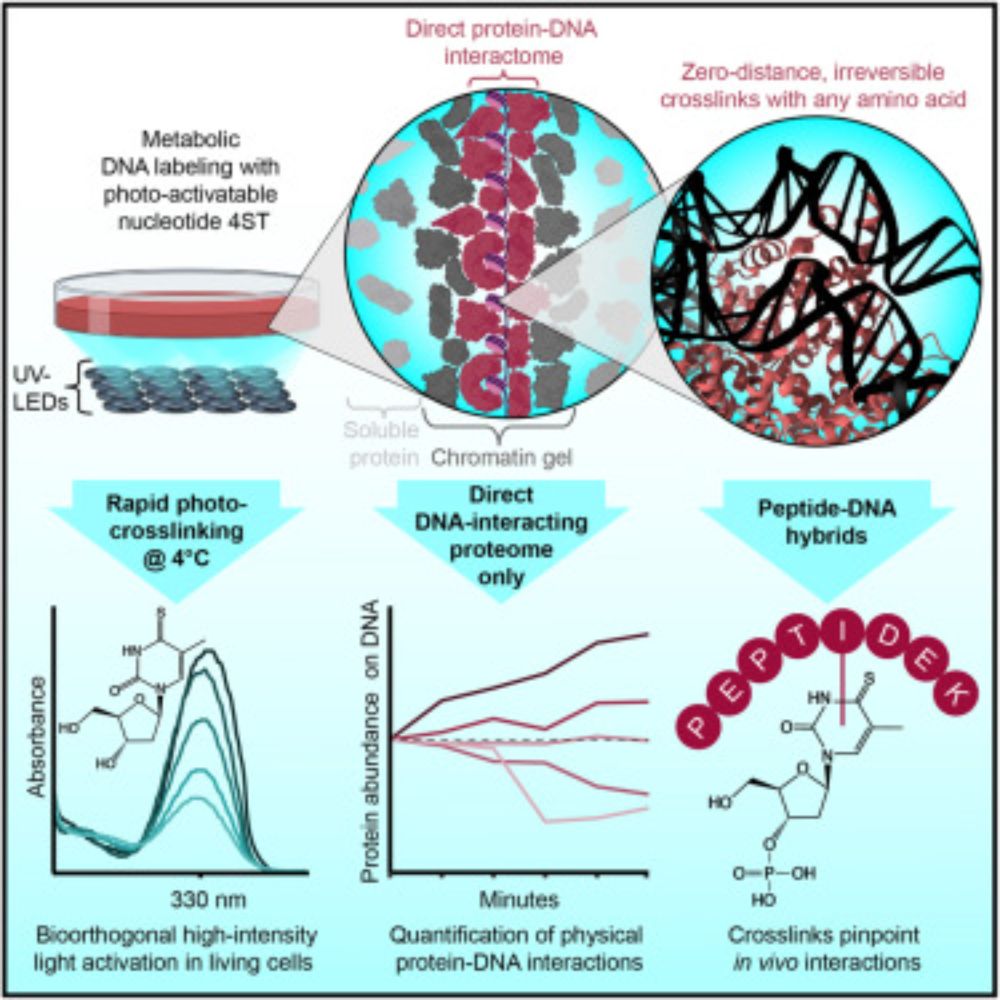
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...

---
#proteomics #prot-other

---
#proteomics #prot-other

---
#proteomics #prot-paper

---
#proteomics #prot-paper
---
#proteomics #prot-paper

---
#proteomics #prot-paper

Sling
ADT
NewDayUSA
AmericaHomeShield
Progressive
LibertyMutual
Shopify
Spectrum
Silac
Quicken
Sterns&Foster
LearCapital
CITI
Pantene
BEHR
Advil
Denver Mattress
Tommie Copper
Invisible Glass
Select Quote
Lexus
SimpliSafe
Sling
ADT
NewDayUSA
AmericaHomeShield
Progressive
LibertyMutual
Shopify
Spectrum
Silac
Quicken
Sterns&Foster
LearCapital
CITI
Pantene
BEHR
Advil
Denver Mattress
Tommie Copper
Invisible Glass
Select Quote
Lexus
SimpliSafe


