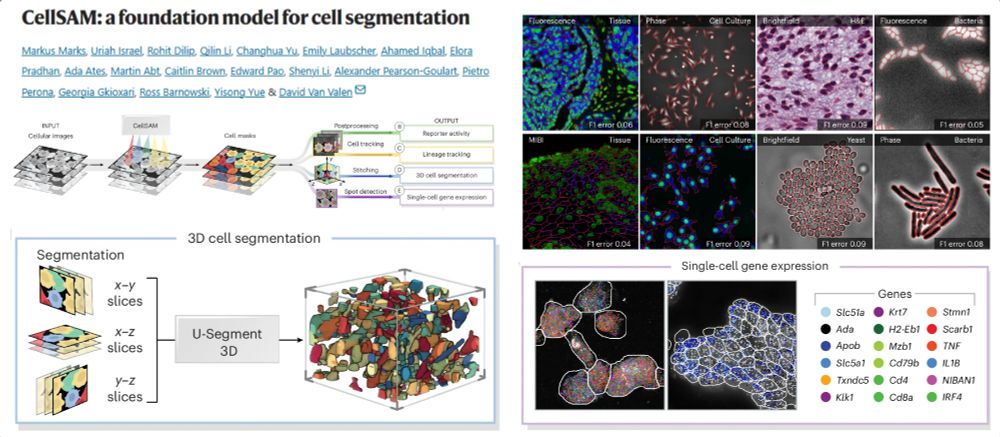
A foundation model for Cell Segmentation
Segment Anything Model - Vision Transformer
cellsam.deepcell.org
Outperforming CellPose
Similar to Human
Single-cell spatial transcriptomics
Live-cell tracking
3D cell segmentation
@natmethods.nature.com 2025
www.nature.com/articles/s41...
A foundation model for Cell Segmentation
Segment Anything Model - Vision Transformer
cellsam.deepcell.org
Outperforming CellPose
Similar to Human
Single-cell spatial transcriptomics
Live-cell tracking
3D cell segmentation
@natmethods.nature.com 2025
www.nature.com/articles/s41...
Check out our preprint: www.biorxiv.org/content/10.1...

Check out our preprint: www.biorxiv.org/content/10.1...
Cellpose 3 is wonderful, easier to install and supports GPUs everywhere.

Cellpose 3 is wonderful, easier to install and supports GPUs everywhere.
There is a dedicated detector for Cellpose SAM. It takes into account the must lower number of parameters. I have included one: the possibility to pass to the model one channel or all channels. This might impact the segmentation accuracy based on your input.
There is a dedicated detector for Cellpose SAM. It takes into account the must lower number of parameters. I have included one: the possibility to pass to the model one channel or all channels. This might impact the segmentation accuracy based on your input.
Cellpose+, a morphological analysis tool for feature extraction of stained cell images
https://arxiv.org/abs/2410.18738
Cellpose+, a morphological analysis tool for feature extraction of stained cell images
https://arxiv.org/abs/2410.18738

Data Efficiency and Transfer Robustness in Biomedical Image Segmentation: A Study of Redundancy and Forgetting with Cellpose
https://arxiv.org/abs/2511.04803
Data Efficiency and Transfer Robustness in Biomedical Image Segmentation: A Study of Redundancy and Forgetting with Cellpose
https://arxiv.org/abs/2511.04803


Why: reproducibility, throughput, objective quantitation
Lore: Reviewers now expect automated or blinded image analysis
(9/10)
Why: reproducibility, throughput, objective quantitation
Lore: Reviewers now expect automated or blinded image analysis
(9/10)
Cycle Hybridization Chain Reaction
14 bp split L+R DNA barcodes
3 color channels
Cellpose-based 3D nucleus segmentation
120 RNA probes + 8 antibodies
~50 µm hippocampal slice
44 imaging cycles over 11 days
@science.org 2025
www.science.org/doi/10.1126/...

Cycle Hybridization Chain Reaction
14 bp split L+R DNA barcodes
3 color channels
Cellpose-based 3D nucleus segmentation
120 RNA probes + 8 antibodies
~50 µm hippocampal slice
44 imaging cycles over 11 days
@science.org 2025
www.science.org/doi/10.1126/...
🔧Setting up Conda environments
🧠Intro to #LLMs in Bioimage Analysis
💡 Mastering Prompt Engineering
🔬 Hands-on with Cellpose for image segmentation
Perfect for bioimage enthusiasts & ML beginners! #AI #Bioinformatics #MachineLearning

🔧Setting up Conda environments
🧠Intro to #LLMs in Bioimage Analysis
💡 Mastering Prompt Engineering
🔬 Hands-on with Cellpose for image segmentation
Perfect for bioimage enthusiasts & ML beginners! #AI #Bioinformatics #MachineLearning


Adapts a foundation model to Cellpose, improving generalization by increasing robustness to image distortions, surpassing inter-human agreement.




Adapts a foundation model to Cellpose, improving generalization by increasing robustness to image distortions, surpassing inter-human agreement.

introduction, we try to understand how the famous CellPose works.
www.nature.com/articles/s41...


introduction, we try to understand how the famous CellPose works.
www.nature.com/articles/s41...
I’m especially thinking of datasets from: CellSeT, Icy (.xml) or .zip ImageJ ROIs (e.g., via Celer, PaCeQuant, Cellpose, etc.)
Something like what’s being done with ggPlantMap would be amazing!
#PlantScience #OpenScience

I’m especially thinking of datasets from: CellSeT, Icy (.xml) or .zip ImageJ ROIs (e.g., via Celer, PaCeQuant, Cellpose, etc.)
Something like what’s being done with ggPlantMap would be amazing!
#PlantScience #OpenScience
paper: www.biorxiv.org/content/10.1...
w/ @computingnature.bsky.social 1/n




