
VS models often learn to predict both cells and noisy background, because training treats all pixels equally. We address this by explicitly training models to prioritize foreground.
1/5


cytodata25.eu-openscreen.eu
cytodata25.eu-openscreen.eu
1/6 #iccv2025
1/6 #iccv2025
The paper: arxiv.org/abs/2503.19545 1/🧵

The paper: arxiv.org/abs/2503.19545 1/🧵
rdcu.be/ezGre
rdcu.be/ezGre

VS models often learn to predict both cells and noisy background, because training treats all pixels equally. We address this by explicitly training models to prioritize foreground.
1/5

VS models often learn to predict both cells and noisy background, because training treats all pixels equally. We address this by explicitly training models to prioritize foreground.
1/5
If you need to obtain interpretable features from your segmented microscopy images, but want to do it in a fully automated way, we know the struggle.
1/6
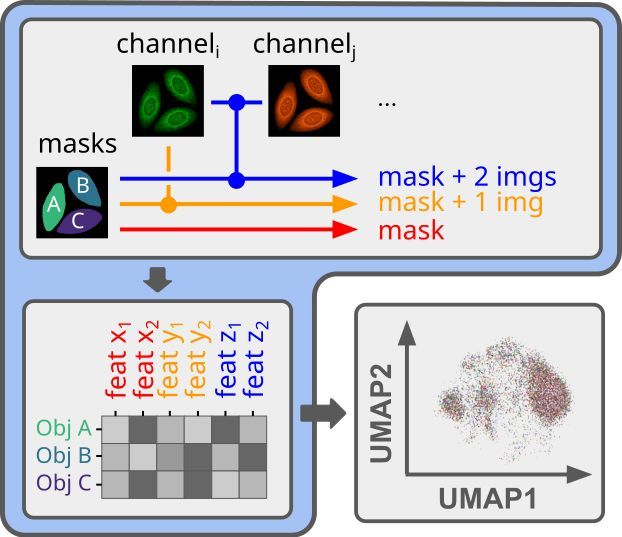
If you need to obtain interpretable features from your segmented microscopy images, but want to do it in a fully automated way, we know the struggle.
1/6
Berlin, November cytodata25.eu-openscreen.eu
Berlin, November cytodata25.eu-openscreen.eu
Calling the AI & bioimaging community to tackle a key microscopy challenge: removing noise while preserving detail.
📦 Paired noisy/clean datasets
📈 Ground-truth evaluation
🧠 DL focus
Build, test, compete 👉 ai4life.eurobioimaging.eu/challenge-2/

Calling the AI & bioimaging community to tackle a key microscopy challenge: removing noise while preserving detail.
📦 Paired noisy/clean datasets
📈 Ground-truth evaluation
🧠 DL focus
Build, test, compete 👉 ai4life.eurobioimaging.eu/challenge-2/
The deadline for abstract submissions for oral presentations at Cytodata 2025 in Berlin is approaching!
👉 𝗦𝘂𝗯𝗺𝗶𝘁 𝘆𝗼𝘂𝗿 𝗮𝗯𝘀𝘁𝗿𝗮𝗰𝘁 𝗯𝘆 𝗝𝘂𝗻𝗲 25!
cytodata25.eu-openscreen.eu/registration/
#BerlinConference #Imageanalysis #Microscopy

www.bioimagecomputing.com

www.bioimagecomputing.com
More in the 🧵 below:
1/7
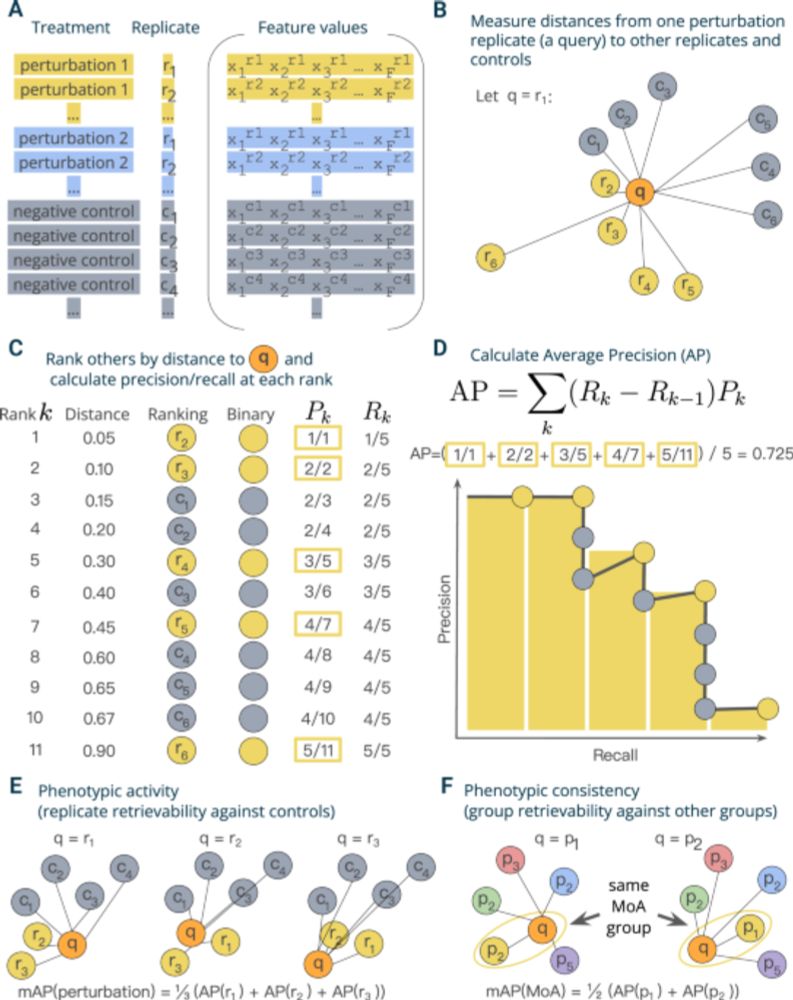
More in the 🧵 below:
1/7
openaccess.thecvf.com/CVPR2025
Workshop papers will be posted shortly. Stay tuned...

openaccess.thecvf.com/CVPR2025
Workshop papers will be posted shortly. Stay tuned...
@biorxivpreprint.bsky.social !!! 😍❤️ biorxiv.org/content/10.1... @scilifelab.se , @crick.ac.uk . More about it in the thread #AI4CellFate (1/n)⬇️

@biorxivpreprint.bsky.social !!! 😍❤️ biorxiv.org/content/10.1... @scilifelab.se , @crick.ac.uk . More about it in the thread #AI4CellFate (1/n)⬇️
👉 Info & registration: cytodata25.eu-openscreen.eu
🔬 Hackathon: Nov 17–19
📢 Symposium: Nov 20–21
Explore advancements in image-based profiling & high-content screening and Tackle real-world challenges using EU-OPENSCREEN data.

paper: www.biorxiv.org/content/10.1...
w/ @computingnature.bsky.social 1/n
We invite North American institutions to submit proposals for the 2026 symposium, whether in-person or hybrid. Nominate your institution by September 1, 2025. Join us in shaping the future of image-based profiling!
docs.google.com/forms/d/e/1F...
We invite North American institutions to submit proposals for the 2026 symposium, whether in-person or hybrid. Nominate your institution by September 1, 2025. Join us in shaping the future of image-based profiling!
docs.google.com/forms/d/e/1F...
github.com/CAREamics/ca...
github.com/CAREamics/ca...
Are there better ways to spend 6 days in mid-July in Boston?Probably.
Would you learn as much as you will here? Probably not.
Apply here:
iac.hms.harvard.edu/bobiac/2025
Are there better ways to spend 6 days in mid-July in Boston?Probably.
Would you learn as much as you will here? Probably not.
Apply here:
iac.hms.harvard.edu/bobiac/2025
An assault on science anywhere is an assault on science everywhere; the global community must unite
🧪
www.nature.com/articles/d41...

An assault on science anywhere is an assault on science everywhere; the global community must unite
🧪
www.nature.com/articles/d41...
Build the best method for segmenting cellular organelles in vEM using our 289 annotated volumes, 40 organelle classes, and 22 diverse cell and tissue types.
🔗 hhmi.news/410evJQ
Build the best method for segmenting cellular organelles in vEM using our 289 annotated volumes, 40 organelle classes, and 22 diverse cell and tissue types.
🔗 hhmi.news/410evJQ

