IGC - University of Edinburgh
Views are my own


I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....

I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
We describe a critical concept that we call *specificity*.
Led by Jeff Spence and Hakhamanesh Mostafavi:
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

We describe a critical concept that we call *specificity*.
Led by Jeff Spence and Hakhamanesh Mostafavi:



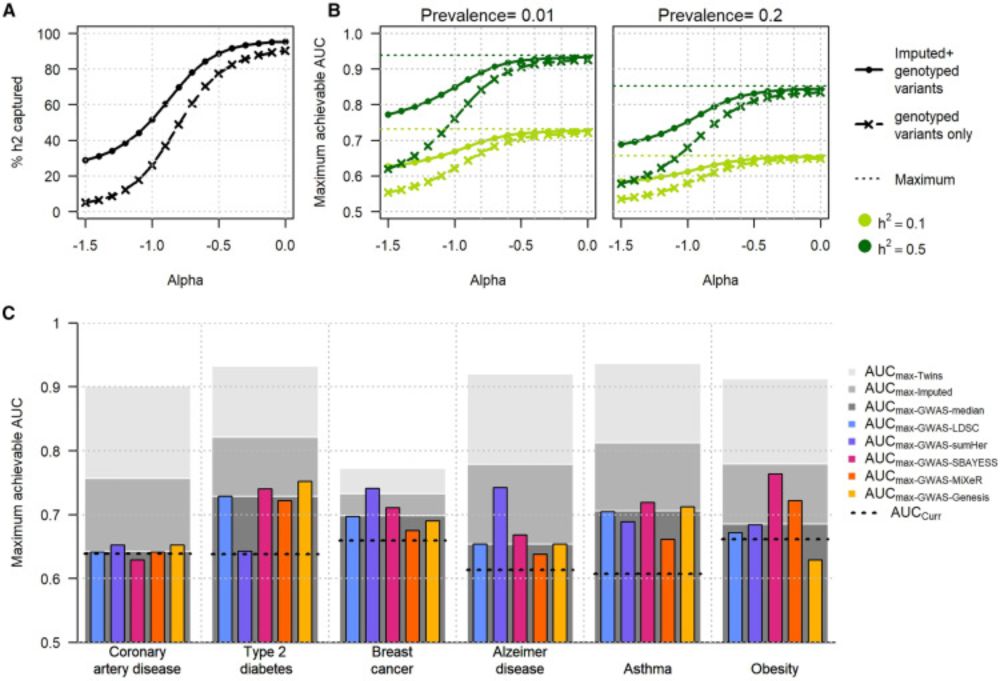
Our new study in Nature Genetics including 9 disease and 7 biobanks shows:
• Susceptibility variants ≠ survival
• PRSs for onset weak at predicting progression
• Lifespan PRS predicts survival better


Our new study in Nature Genetics including 9 disease and 7 biobanks shows:
• Susceptibility variants ≠ survival
• PRSs for onset weak at predicting progression
• Lifespan PRS predicts survival better
My love letter to social science genetics: communities.springernature.com/posts/a-love...

My love letter to social science genetics: communities.springernature.com/posts/a-love...
Populations differ in traits/disease burden. Are these differences due to genetics?
Comparing single variants or polygenic scores between populations is biased due to environmental confounders correlated with the variants.
1/3
www.medrxiv.org/content/10.1...

Populations differ in traits/disease burden. Are these differences due to genetics?
Comparing single variants or polygenic scores between populations is biased due to environmental confounders correlated with the variants.
1/3
www.medrxiv.org/content/10.1...

Here we share our FLAMES framework, which predicts the effector genes in GWAS loci with state-of-the-art precision🔥
Special thanks to @daniposthu.bsky.social
A full thread describing findings below!
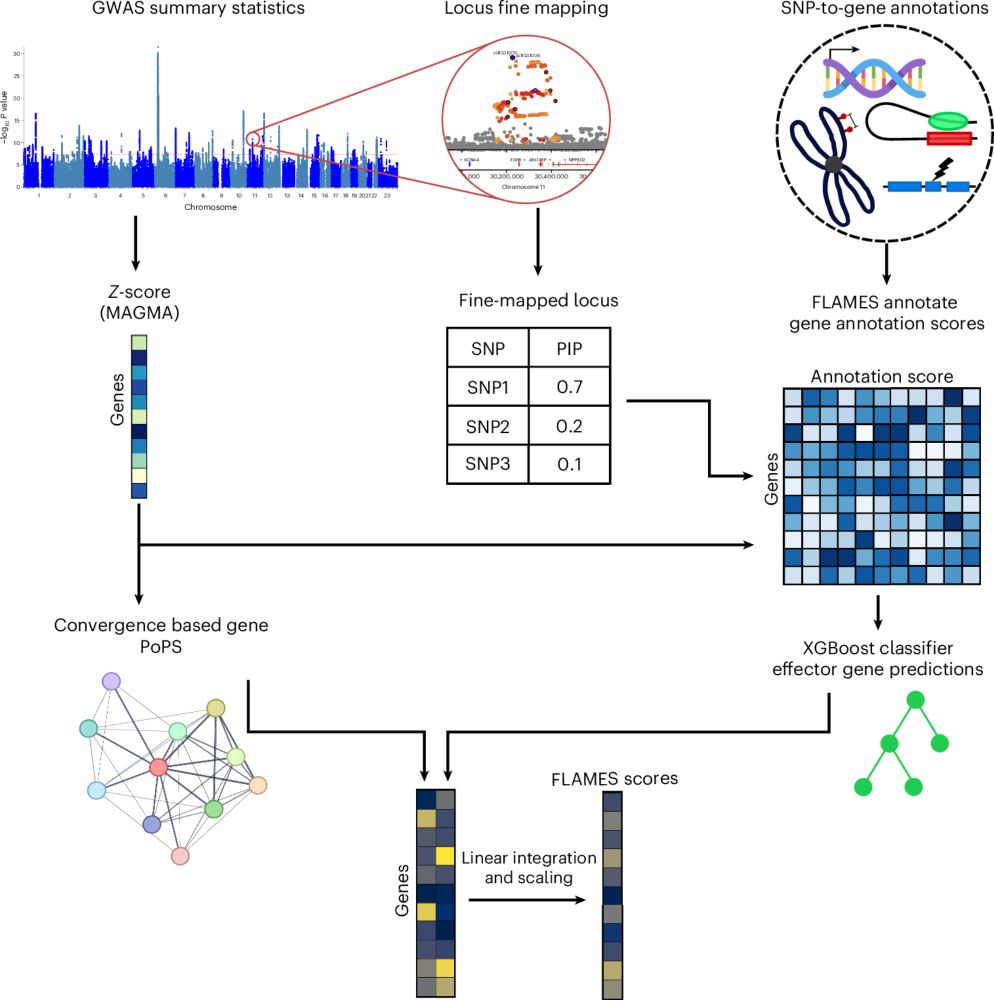
Here we share our FLAMES framework, which predicts the effector genes in GWAS loci with state-of-the-art precision🔥
Special thanks to @daniposthu.bsky.social
A full thread describing findings below!
- our new tool FLAMES 🔥🔥, to identify the most likely causal genes in GWAS loci, using machine learning techniques, thoroughly benchmarked and freely available
Paper: rdcu.be/d9iQP
Code: github.com/Marijn-Schip...

- our new tool FLAMES 🔥🔥, to identify the most likely causal genes in GWAS loci, using machine learning techniques, thoroughly benchmarked and freely available
Paper: rdcu.be/d9iQP
Code: github.com/Marijn-Schip...
link: rdcu.be/ezuZd
1/7

link: rdcu.be/ezuZd
1/7
Higher eQTL power reveals signals that boost GWAS colocalization 🧪🧬
www.biorxiv.org/content/10.1...

Higher eQTL power reveals signals that boost GWAS colocalization 🧪🧬
www.biorxiv.org/content/10.1...
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
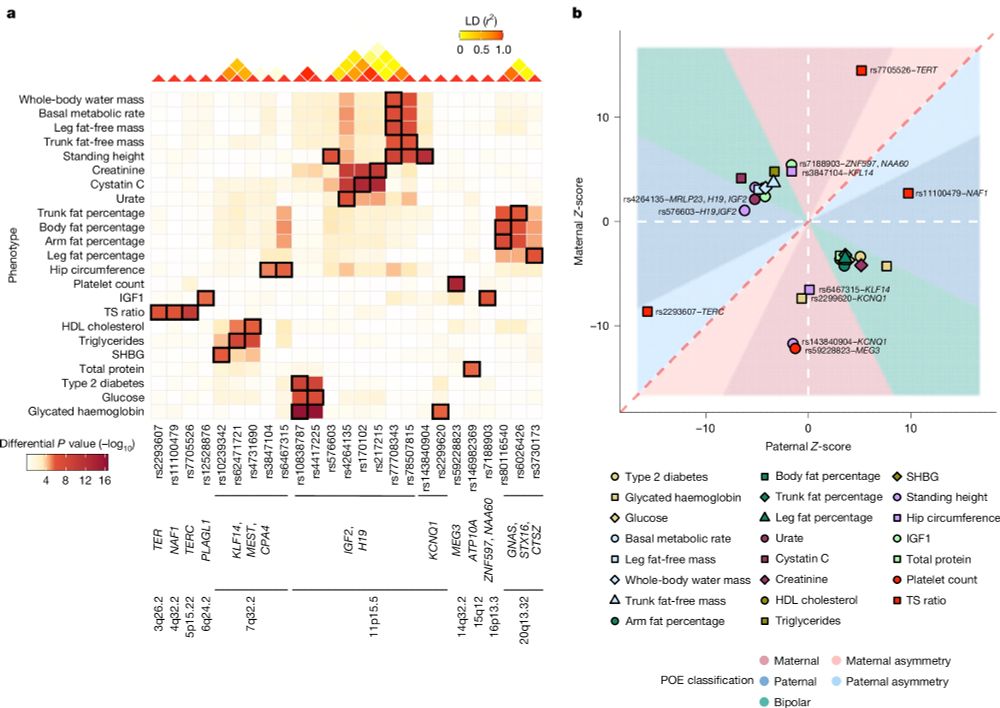
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
Read here: www.nature.com/articles/s41...
Read here: www.nature.com/articles/s41...
Another story of consent for genetic data use this week.
www.science.org/content/arti...

Another story of consent for genetic data use this week.
www.mv.helsinki.fi/home/mjxpiri...
www.mv.helsinki.fi/home/mjxpiri...
📄Haplotype analysis reveals pleiotropic disease associations in the HLA region
🧑🤝🧑 @jkpritch.bsky.social @finngen.bsky.social & colleagues