
Tim Straub
@timothystraub.com
Senior Data Scientist @ Tiny Health
Data science | bioinformatics| computational biology
Broadly interested in NGS and multi-omics, microbiome, infectious disease, ML/AI, cool science, and big data. Also golden retrievers.
All views are my own.
Data science | bioinformatics| computational biology
Broadly interested in NGS and multi-omics, microbiome, infectious disease, ML/AI, cool science, and big data. Also golden retrievers.
All views are my own.
Pinned
Tim Straub
@timothystraub.com
· Sep 4

Improving immune‐related health outcomes post‐cesarean birth with a gut microbiome‐based program: A randomized controlled trial
Background Infants born via Cesarean section (C-section) often have a distinct gut microbiome and higher risks of atopic and immune-related conditions than vaginally delivered infants. We evaluated ...
onlinelibrary.wiley.com
1/3 🧬 Exciting news from my colleagues at Tiny Health!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!
Reposted by Tim Straub
Can we leverage bacterial competition for targeted replacement of harmful strains? Maybe! Our recent piece in @natmicrobiol.nature.com provides a theoretical framework and a set of experiments to show what it might take: www.nature.com/articles/s41...

Strain displacement in microbiomes via ecological competition - Nature Microbiology
Mathematical modelling and experimental tests reveal principles that govern displacement of a resident strain by an invader in microbial communities.
www.nature.com
November 7, 2025 at 10:34 PM
Can we leverage bacterial competition for targeted replacement of harmful strains? Maybe! Our recent piece in @natmicrobiol.nature.com provides a theoretical framework and a set of experiments to show what it might take: www.nature.com/articles/s41...
Reposted by Tim Straub
Interested in #drugs, #microbiome and #antibiotic resistance, then check out our paper on the effects of over 300 drugs on the gut microbiome. Exciting results as well on the role of antibiotic resistance defining how microbiomes respond to drugs.
@quadraminstitute.bsky.social
rdcu.be/eMmcT
@quadraminstitute.bsky.social
rdcu.be/eMmcT

Systematic metaproteomics mapping reveals functional and ecological landscapes of Ex vivo human gut microbiota responses to therapeutic drugs
Nature Communications - Here, the authors systematically map metaproteomic responses of ex vivo human gut microbiota to common therapeutics, identifying several drug classes inducing strong...
rdcu.be
October 24, 2025 at 4:20 PM
Interested in #drugs, #microbiome and #antibiotic resistance, then check out our paper on the effects of over 300 drugs on the gut microbiome. Exciting results as well on the role of antibiotic resistance defining how microbiomes respond to drugs.
@quadraminstitute.bsky.social
rdcu.be/eMmcT
@quadraminstitute.bsky.social
rdcu.be/eMmcT
Reposted by Tim Straub
Looks interesting 👀: Distinguishing diet- and microbe-derived metabolites in the human gut microbiomejournal.biomedcentral.com/articles/10....

Distinguishing diet- and microbe-derived metabolites in the human gut - Microbiome
Background Human gut microbes metabolize food and host secretions, consuming and producing small molecules that are important to health and homeostasis. Here, we present an atlas of diet- and microbio...
microbiomejournal.biomedcentral.com
October 18, 2025 at 7:07 PM
Looks interesting 👀: Distinguishing diet- and microbe-derived metabolites in the human gut microbiomejournal.biomedcentral.com/articles/10....
I welcome another promising tool to analyze strain-resolved WMS data! #microbiome
Strainify: Strain-Level Microbiome Profiling for Low-Coverage Short-Read Metagenomic Datasets https://www.biorxiv.org/content/10.1101/2025.10.10.681738v1
October 14, 2025 at 1:59 AM
I welcome another promising tool to analyze strain-resolved WMS data! #microbiome
👀 What an interesting and under-explored idea!
Is a healthy microbiome one that is rich in phages? 🦠 Excited to share our paper out in Lancet Microbe with @bkoskella.bsky.social & @dholtappels.bsky.social where we test whether virome diversity can be used a broad signature of microbiome health 📈
New research article
Evaluation of bacteriophages as a signature of #microbiome health: a systematic review and meta-analysis
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #ViroSky #Phage #OpenAccess #OA
Evaluation of bacteriophages as a signature of #microbiome health: a systematic review and meta-analysis
www.thelancet.com/journals/lan...
#IDSky #ClinMicro #ViroSky #Phage #OpenAccess #OA
October 10, 2025 at 2:39 PM
👀 What an interesting and under-explored idea!
Reposted by Tim Straub
SAVE THE DATE: 27 October, 2025
and join speakers from both illumina and #MVIF!
Registration for the webinar is free: emea.illumina.com/events/webin...
and join speakers from both illumina and #MVIF!
Registration for the webinar is free: emea.illumina.com/events/webin...

October 10, 2025 at 1:58 PM
SAVE THE DATE: 27 October, 2025
and join speakers from both illumina and #MVIF!
Registration for the webinar is free: emea.illumina.com/events/webin...
and join speakers from both illumina and #MVIF!
Registration for the webinar is free: emea.illumina.com/events/webin...
👀 One of the first two articles published for the collection on #Microbiome and Reproductive Health!
link.springer.com/collections/...
link.springer.com/collections/...
October 3, 2025 at 3:56 PM
👀 One of the first two articles published for the collection on #Microbiome and Reproductive Health!
link.springer.com/collections/...
link.springer.com/collections/...
Reposted by Tim Straub
Happy to share this paper in final form rdcu.be/eH8tj, with more info on neuronal responses and potential mechanism of actions! The results suggest that there is neural interoception of microbial metabolic state 🧠🦠 We hope they can inspire more work in this area!
September 25, 2025 at 3:36 PM
Happy to share this paper in final form rdcu.be/eH8tj, with more info on neuronal responses and potential mechanism of actions! The results suggest that there is neural interoception of microbial metabolic state 🧠🦠 We hope they can inspire more work in this area!
Reposted by Tim Straub
our software, microbetag, for the annotation of microbial co-occurrence networks with phenotypic traits & metabolic complementarities was just published
A Cytoscape app is also available to make your life easier (and prettier)
#microbiome #metabolic-modeling #networks
A Cytoscape app is also available to make your life easier (and prettier)
#microbiome #metabolic-modeling #networks

microbetag: simplifying microbial network interpretation through annotation, enrichment tests, and metabolic complementarity analysis - Genome Biology
Microbial co-occurrence network inference is often hindered by low accuracy and tool dependency. We introduce microbetag, a comprehensive software ecosystem designed to annotate microbial networks. No...
doi.org
September 23, 2025 at 9:42 AM
our software, microbetag, for the annotation of microbial co-occurrence networks with phenotypic traits & metabolic complementarities was just published
A Cytoscape app is also available to make your life easier (and prettier)
#microbiome #metabolic-modeling #networks
A Cytoscape app is also available to make your life easier (and prettier)
#microbiome #metabolic-modeling #networks
👀 This looks really interesting–being able to understand genomic context and assess species carriage of #ARGs from WMS data. #microbiome
CARD k-mers: Unmasking the pathogen hosts and genomic contexts of antimicrobial resistance genes in metagenomic sequences https://www.biorxiv.org/content/10.1101/2025.09.15.676352v1
September 19, 2025 at 2:26 AM
👀 This looks really interesting–being able to understand genomic context and assess species carriage of #ARGs from WMS data. #microbiome
Reposted by Tim Straub
We are looking for a postdoc to study single-cell transcriptional heterogeneity in the human skin microbiome.
We have a new protocol mostly developed, but need someone to see it through. Experience with protocol dev or RNAseq appreciated.
Funded by a new grant from MIT-HEALS.
We have a new protocol mostly developed, but need someone to see it through. Experience with protocol dev or RNAseq appreciated.
Funded by a new grant from MIT-HEALS.
September 12, 2025 at 4:49 PM
We are looking for a postdoc to study single-cell transcriptional heterogeneity in the human skin microbiome.
We have a new protocol mostly developed, but need someone to see it through. Experience with protocol dev or RNAseq appreciated.
Funded by a new grant from MIT-HEALS.
We have a new protocol mostly developed, but need someone to see it through. Experience with protocol dev or RNAseq appreciated.
Funded by a new grant from MIT-HEALS.
🧬 Research from the American Gut Project shows that people who eat 30+ different plants per week tend to have a more diverse gut #microbiome than those eating under 10.
At Tiny Health, we're running a fun 7-day challenge to help you get there—plus one winner gets a gut health test ($199 value)!
At Tiny Health, we're running a fun 7-day challenge to help you get there—plus one winner gets a gut health test ($199 value)!

Enter Tiny Health's 30 Plants Challenge for your chance to win big
Join Tiny Health’s free 7-day gut health challenge and get expert tips, daily inspiration, and recipes to help you reach 30 different plants and a healthier gut. Why 30? Research shows that eating a w...
www.tinyhealth.com
September 12, 2025 at 5:15 PM
🧬 Research from the American Gut Project shows that people who eat 30+ different plants per week tend to have a more diverse gut #microbiome than those eating under 10.
At Tiny Health, we're running a fun 7-day challenge to help you get there—plus one winner gets a gut health test ($199 value)!
At Tiny Health, we're running a fun 7-day challenge to help you get there—plus one winner gets a gut health test ($199 value)!
Reposted by Tim Straub
MAGdb: a comprehensive high quality MAGs repository for exploring microbial metagenome-assemble genomes genomebiology.biomedcentral.com/articles/10.... #jcampubs

MAGdb: a comprehensive high quality MAGs repository for exploring microbial metagenome-assemble genomes - Genome Biology
Metagenomic analyses of microbial communities have unveiled a substantial level of interspecies and intraspecies genetic diversity by reconstructing metagenome-assembled genomes (MAGs). The MAG databa...
genomebiology.biomedcentral.com
September 11, 2025 at 4:03 PM
MAGdb: a comprehensive high quality MAGs repository for exploring microbial metagenome-assemble genomes genomebiology.biomedcentral.com/articles/10.... #jcampubs
Authors argue that integrating metagenomics, culturing, and experimental validation to study hidden yet ecologically significant microbial species will advance our understanding of the microbiome in health and disease, and inform next-gen therapeutics leveraging the full diversity of the gut.
🚨 New pre-print from the lab! We performed a large-scale meta-analysis of the uncultured gut #microbiome in >10,000 metagenomes enabling us to identify a new candidate biomarker of health. www.biorxiv.org/content/10.1...

Meta-analysis of the uncultured gut microbiome across 11,115 global metagenomes reveals a new candidate biomarker of health
The human gut microbiome plays an important biological role in host health, yet over 60% of gut species remain uncultured and hence inaccessible to experimental manipulation. Here we analysed 11,115 h...
www.biorxiv.org
September 10, 2025 at 1:11 PM
Authors argue that integrating metagenomics, culturing, and experimental validation to study hidden yet ecologically significant microbial species will advance our understanding of the microbiome in health and disease, and inform next-gen therapeutics leveraging the full diversity of the gut.
Reposted by Tim Straub
There are two recent papers that use sourmash in creative ways - and it gladdens my heart! pubmed.ncbi.nlm.nih.gov/40812187/ by @silask.bsky.social et al looks cleverly at human gut subspecies. www.biorxiv.org/content/10.1... by @rayanchikhi.bsky.social et al counts billions of k-mers. Super neat!

Subspecies of the human gut microbiota carry implicit information for in-depth microbiome research - PubMed
Microbial strains within a single species can exhibit distinct functional characteristics due to variations in gene content and often show individual specificity, which can obscure unbiased associatio...
pubmed.ncbi.nlm.nih.gov
September 8, 2025 at 1:49 PM
There are two recent papers that use sourmash in creative ways - and it gladdens my heart! pubmed.ncbi.nlm.nih.gov/40812187/ by @silask.bsky.social et al looks cleverly at human gut subspecies. www.biorxiv.org/content/10.1... by @rayanchikhi.bsky.social et al counts billions of k-mers. Super neat!
Reposted by Tim Straub
A Nature Outlook article reports on the preliminary evidence that microbiota-based treatments can be beneficial for people with mood disorders, such as depression and anxiety. But how these interventions work is not clear, and human trials are needed. #microbiome #medsky 🧪

Why nurturing the gut microbiota could resolve depression and anxiety
Links between gut microbes and mental health could lead to large-scale trials of probiotic interventions.
go.nature.com
September 6, 2025 at 1:31 AM
A Nature Outlook article reports on the preliminary evidence that microbiota-based treatments can be beneficial for people with mood disorders, such as depression and anxiety. But how these interventions work is not clear, and human trials are needed. #microbiome #medsky 🧪
1/3 🧬 Exciting news from my colleagues at Tiny Health!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!

Improving immune‐related health outcomes post‐cesarean birth with a gut microbiome‐based program: A randomized controlled trial
Background Infants born via Cesarean section (C-section) often have a distinct gut microbiome and higher risks of atopic and immune-related conditions than vaginally delivered infants. We evaluated ...
onlinelibrary.wiley.com
September 4, 2025 at 12:51 AM
1/3 🧬 Exciting news from my colleagues at Tiny Health!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!
New study: A personalized #microbiome program helped C-section babies develop more normal gut microbiomes AND showed 83% lower odds of atopic dermatitis!
Just came across this preprint: groups at @broadinstitute.org and MGH developed a low-cost, #CRISPR based diagnostic for pathogenic bacteria and #AMR genes from blood cultures.
BADLOCK: A Rapid, Portable, Inexpensive Diagnostic for Bacterial Pathogen and Resistance Detection in Resource-Limited Settings https://www.medrxiv.org/content/10.1101/2025.08.11.25332217v1
August 31, 2025 at 1:07 AM
Just came across this preprint: groups at @broadinstitute.org and MGH developed a low-cost, #CRISPR based diagnostic for pathogenic bacteria and #AMR genes from blood cultures.
Reposted by Tim Straub
The potential relevance of FMT as a #microbiome augmenting intervention for #obesity management and metabolic health
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Long-term health outcomes in adolescents with obesity treated with faecal microbiota transplantation: 4-year follow-up - Nature Communications
In this 4-year unblinded follow-up study, the authors evaluate 63% of the participants of a double-blind randomised control trial that assessed the effects of faecal microbiota transplantation (FMT) o...
www.nature.com
August 28, 2025 at 9:58 AM
The potential relevance of FMT as a #microbiome augmenting intervention for #obesity management and metabolic health
www.nature.com/articles/s41...
www.nature.com/articles/s41...
This looks really promising! It’s a step in the right direction to drill down to a more function based microbiome analysis while still maintaining taxonomic information.
New paper out: Subspecies of the human gut microbiota carry implicit information for in-depth microbiome research.

August 29, 2025 at 10:03 PM
This looks really promising! It’s a step in the right direction to drill down to a more function based microbiome analysis while still maintaining taxonomic information.
Reposted by Tim Straub
Congrats to postdoc Zhe Zhou on her paper out today in @cp-cellhostmicrobe.bsky.social showing that gut bacteria cross-feed a common dietary antioxidant to produce energy under anaerobic conditions. Thanks to collaborators Angela Jiang and Xiaofang Jiang at NIH.
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
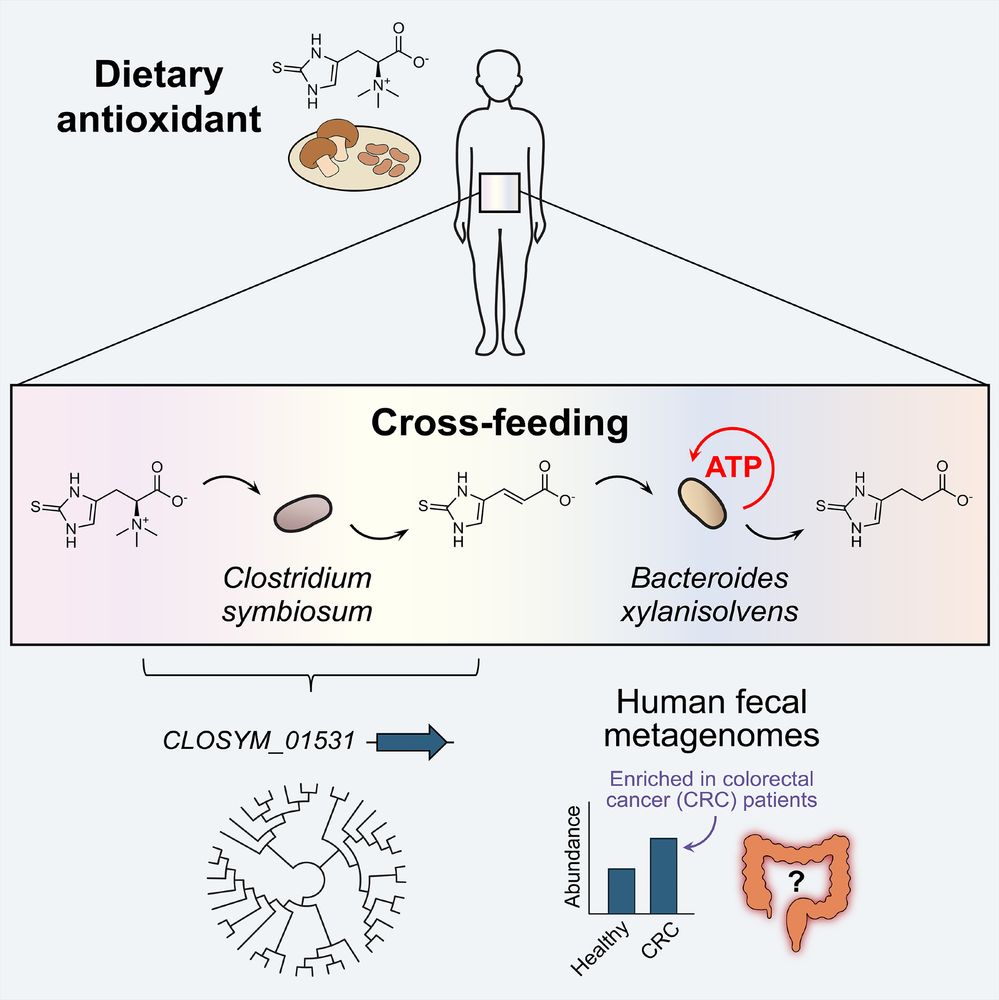
August 4, 2025 at 6:55 PM
Congrats to postdoc Zhe Zhou on her paper out today in @cp-cellhostmicrobe.bsky.social showing that gut bacteria cross-feed a common dietary antioxidant to produce energy under anaerobic conditions. Thanks to collaborators Angela Jiang and Xiaofang Jiang at NIH.
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
Reposted by Tim Straub
Is the gut microbiota composition in the 1st week of life linked to viral lower respiratory tract infections (vLRTIs) in early childhood? Here, work using the BBS birth cohort identifies Bifidobacterium longum-dominated cluster linked to lowest vLRTI rates
🔗 www.sciencedirect.com/science/arti...
🔗 www.sciencedirect.com/science/arti...

Investigation of associations between the neonatal gut microbiota and severe viral lower respiratory tract infections in the first 2 years of life: a birth cohort study with metagenomics
Early-life gut microbiota affects immune system development, including the lung immune response (gut–lung axis). We aimed to investigate whether gut m…
www.sciencedirect.com
August 5, 2025 at 8:33 PM
Is the gut microbiota composition in the 1st week of life linked to viral lower respiratory tract infections (vLRTIs) in early childhood? Here, work using the BBS birth cohort identifies Bifidobacterium longum-dominated cluster linked to lowest vLRTI rates
🔗 www.sciencedirect.com/science/arti...
🔗 www.sciencedirect.com/science/arti...
This study published today finds "a more diverse gut microbiome is generally associated with healthier glucose spike metrics." It's certainly not implying causality, but it is very interesting to see the association!
Our @naturemedicine.bsky.social paper on glucose spikes (CGM sensor) and their relationship with physical activity, gut microbiome, genomics, food intake, and sleep via multimodal A.I. Helps identify people with prediabetes who have high risk of progression (cf HbA1c)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
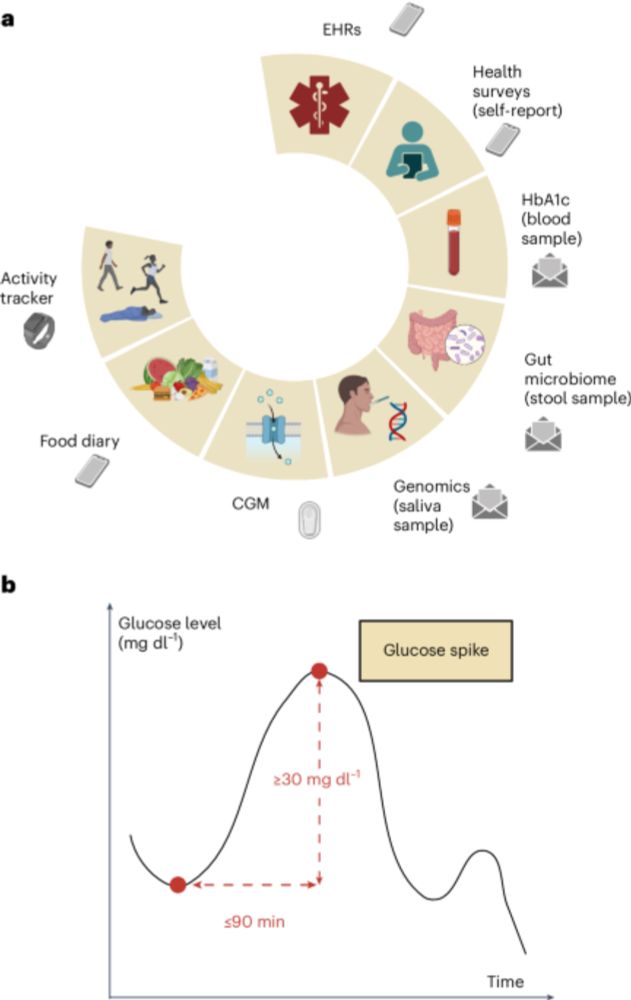
Multimodal AI correlates of glucose spikes in people with normal glucose regulation, pre-diabetes and type 2 diabetes - Nature Medicine
Multimodal data from 347 deeply phenotyped individuals including healthy, prediabetic individuals and individuals with T2D report remotely acquired patterns of glucose control via continuous glucose m...
www.nature.com
July 31, 2025 at 1:28 PM
This study published today finds "a more diverse gut microbiome is generally associated with healthier glucose spike metrics." It's certainly not implying causality, but it is very interesting to see the association!
Reposted by Tim Straub
#WeekendRead! #EveryCellIsAnImmuneCell! #TollPower! Kaelberer Bohorquez &co show @nature.com that #TLR5 signaling in response to flagellin drives neuropeptide PYY release by gut colonic neuropod cells, activating vagal neurons that signal to #brain, reducing feeding via a “neurobiotic sense”!

A gut sense for a microbial pattern regulates feeding - Nature
A study reveals a gut–brain sensory pathway through which the microbial component flagellin activates neuropod cells in the colon to signal the brain and reduce feeding in mice.
www.nature.com
July 26, 2025 at 3:59 PM
#WeekendRead! #EveryCellIsAnImmuneCell! #TollPower! Kaelberer Bohorquez &co show @nature.com that #TLR5 signaling in response to flagellin drives neuropeptide PYY release by gut colonic neuropod cells, activating vagal neurons that signal to #brain, reducing feeding via a “neurobiotic sense”!
Reposted by Tim Straub
The Lynch lab’s latest research delves into the gut microbiome’s influence on peanut oral immunotherapy outcomes. The study identifies specific bile acids and microbial metabolic signatures that predict treatment success. #Microbiome #AllergyResearch #UCSF rdcu.be/ewbEn
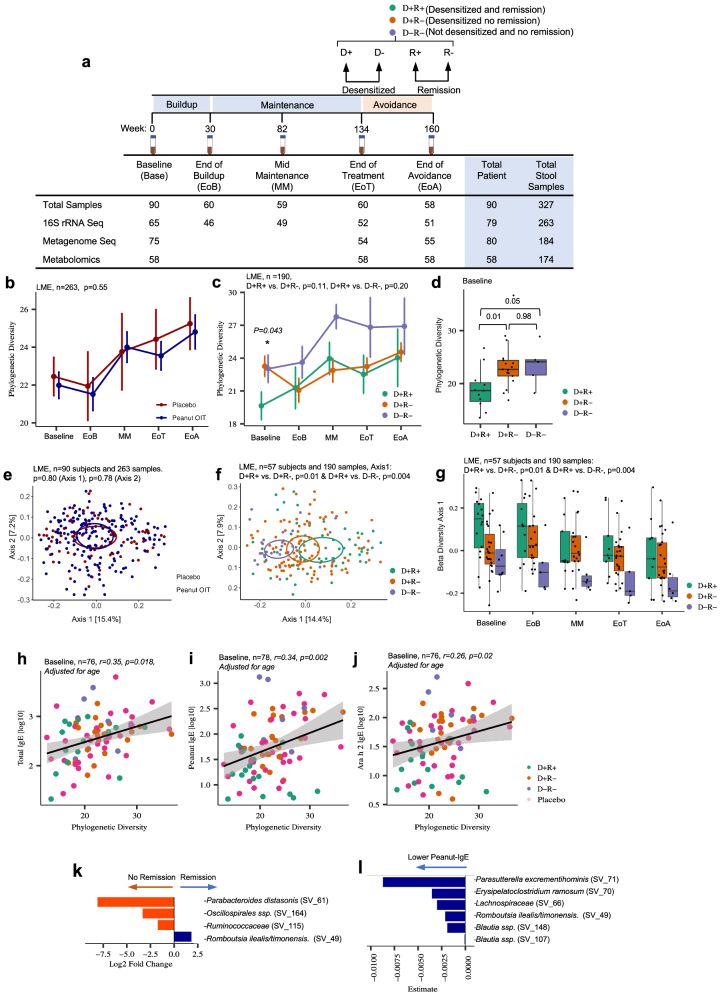
Gut microbial bile and amino acid metabolism associate with peanut oral immunotherapy failure
Nature Communications - Peanut oral immunotherapy (POIT) can treat peanut allergy, but only a subset of patients achieve lasting remission. Here, the authors show that POIT efficacy is associated...
rdcu.be
July 14, 2025 at 3:36 PM
The Lynch lab’s latest research delves into the gut microbiome’s influence on peanut oral immunotherapy outcomes. The study identifies specific bile acids and microbial metabolic signatures that predict treatment success. #Microbiome #AllergyResearch #UCSF rdcu.be/ewbEn

