Thomas Balan
@thomasbalan.bsky.social
Postdoc in the Duharcourt lab at the Jacques Monod Institute
Sudying histone modifications during programmed DNA elimination in Paramecium
PhD from @upcite.bsky.social @ijmonod.bsky.social
Sudying histone modifications during programmed DNA elimination in Paramecium
PhD from @upcite.bsky.social @ijmonod.bsky.social
Reposted by Thomas Balan
Excited to share our new preprint on BioRxiv!
A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5
A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5

The tiny germline chromosomes of Paramecium aurelia have an exceptionally high recombination rate and are capped by a new class of Helitrons
Background. Paramecia belong to the ciliate phylum of unicellular eukaryotes characterized by nuclear dimorphism. A diploid germline micronucleus (MIC) transmits genetic information across sexual gene...
www.biorxiv.org
November 10, 2025 at 9:21 AM
Excited to share our new preprint on BioRxiv!
A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5
A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5
Reposted by Thomas Balan
Lab’s first paper is out!! We show the first structures of #Asgard #chromatin by #cryo-EM 🧬❄️
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

October 28, 2025 at 3:07 PM
Lab’s first paper is out!! We show the first structures of #Asgard #chromatin by #cryo-EM 🧬❄️
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Reposted by Thomas Balan
How do pleiotropic TFs generate organized diversity in developing tissues? @spinalorga.bsky.social shows that PAX3 & PAX7 organize #SpinalCord by acting as both repressors & pioneer activators, regulated by #morphogens to ensure precise neural subtype specification @plosbiology.org 🧪 plos.io/4qumsla

October 27, 2025 at 5:25 PM
How do pleiotropic TFs generate organized diversity in developing tissues? @spinalorga.bsky.social shows that PAX3 & PAX7 organize #SpinalCord by acting as both repressors & pioneer activators, regulated by #morphogens to ensure precise neural subtype specification @plosbiology.org 🧪 plos.io/4qumsla
🚨 Excited to share my first preprint on bioRxiv! 🎓 We uncover a non-canonical role of condensin I, beyond chromosome segregation 🧬
This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...
This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...

A developmental condensin I complex assists the Paramecium PiggyMac domesticated transposase during programmed DNA elimination
Prokaryotes and eukaryotes use diverse strategies to cope with invading mobile genetic elements, including programmed DNA elimination (PDE). In the ciliate Paramecium , elimination of transposable ele...
doi.org
October 21, 2025 at 2:46 PM
🚨 Excited to share my first preprint on bioRxiv! 🎓 We uncover a non-canonical role of condensin I, beyond chromosome segregation 🧬
This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...
This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...
Reposted by Thomas Balan
Our latest paper on a histone methyltransferase-independent function of PRC2 controlling small RNA dynamics during programmed DNA elimination in Paramecium is now published in #NAR.
#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...
#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...

A histone methyltransferase-independent function of PRC2 controls small RNA dynamics during programmed DNA elimination in Paramecium
Abstract. To limit transposable element (TE) mobilization, most eukaryotes have evolved small RNAs to silence TE activity via homology-dependent mechanisms
academic.oup.com
October 17, 2025 at 11:58 AM
Our latest paper on a histone methyltransferase-independent function of PRC2 controlling small RNA dynamics during programmed DNA elimination in Paramecium is now published in #NAR.
#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...
#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...
Reposted by Thomas Balan
Huge congratulations to Thomas on this well-deserved award. His PhD defense last week was outstanding too - a great moment. He’ll on the lookout for a postdoc position soon…spread the word!
📣 Congratulations to @thomasbalan.bsky.social (@sandraduharcourt.bsky.social Lab) who won the Best Poster Award at the Gordon Research Epigenetics conference “Variation: Mechanisms and Impact across Systems”!
More informations 🔗 buff.ly/NhLWE6G
@cnrs-idf-villejuif.bsky.social @upcite.bsky.social
More informations 🔗 buff.ly/NhLWE6G
@cnrs-idf-villejuif.bsky.social @upcite.bsky.social

September 23, 2025 at 3:25 PM
Huge congratulations to Thomas on this well-deserved award. His PhD defense last week was outstanding too - a great moment. He’ll on the lookout for a postdoc position soon…spread the word!
Reposted by Thomas Balan
Please re-post:
Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September
Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September

EvoChromo: Evolutionary approaches to research in chromatin
Chromatin is the complex of DNA, RNA and protein that is found making up the chromosomes in eukaryotic cells. Chromatin is essential for proper genome function and is involved in chromosome segregati…
meetings.embo.org
September 8, 2025 at 11:14 AM
Please re-post:
Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September
Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September
👀
📢 PhD Thesis defense
📅 Sept. 19th
📍 IJM
@thomasbalan.bsky.social ( @sandraduharcourt.bsky.social Lab ) will defend his PhD thesis "Characterization of proteins involved in the dynamics of histone modifications during programmed DNA elimination in Paramecium tetraurelia"
➡️ buff.ly/V8vzSsT
📅 Sept. 19th
📍 IJM
@thomasbalan.bsky.social ( @sandraduharcourt.bsky.social Lab ) will defend his PhD thesis "Characterization of proteins involved in the dynamics of histone modifications during programmed DNA elimination in Paramecium tetraurelia"
➡️ buff.ly/V8vzSsT

August 26, 2025 at 12:15 PM
👀
Reposted by Thomas Balan
📢 Institut Jacques Monod Seminar
📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ
📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ

August 25, 2025 at 12:02 PM
📢 Institut Jacques Monod Seminar
📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ
📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ
Reposted by Thomas Balan
New online: Binding of heterochromatin protein Rhino to a subset of piRNA clusters depends on a combination of two histone marks

Binding of heterochromatin protein Rhino to a subset of piRNA clusters depends on a combination of two histone marks
Nature Structural & Molecular Biology, Published online: 17 June 2025; doi:10.1038/s41594-025-01584-8The authors reveal that, in fruit fly ovaries, the protein Rhino is guided to specific regions of the genome by a combination of two histone modifications, enhancing understanding of how cells protect their DNA from harmful virus-like elements.
go.nature.com
June 17, 2025 at 10:29 AM
New online: Binding of heterochromatin protein Rhino to a subset of piRNA clusters depends on a combination of two histone marks
Reposted by Thomas Balan
Mechanisms and timing of programmed DNA elimination in songbirds www.biorxiv.org/content/10.1...

Mechanisms and timing of programmed DNA elimination in songbirds
It is commonly assumed that multicellular organisms contain the same genetic information in all the cells of an individual. However, there is a growing list of species in which parts of the genome are...
www.biorxiv.org
June 1, 2025 at 9:24 PM
Mechanisms and timing of programmed DNA elimination in songbirds www.biorxiv.org/content/10.1...
Reposted by Thomas Balan
We’ve uncovered Asgard chromatin structures formed by a Hodarchaeal histone : closed hypernucleosome conserved in archaea and an open form resembling the H3-H4 eukaryotic octasome. Fantastic work by @harshranawat.bsky.social!
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
May 26, 2025 at 4:56 PM
We’ve uncovered Asgard chromatin structures formed by a Hodarchaeal histone : closed hypernucleosome conserved in archaea and an open form resembling the H3-H4 eukaryotic octasome. Fantastic work by @harshranawat.bsky.social!
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
I add an amazing time last week at the Epigenetics 2025 meeting from the @gensocuk.bsky.social in sunny Belfast! Great discussions and insights on my PhD project! Also very honored to have been awarded the best poster prize!

April 30, 2025 at 11:23 AM
I add an amazing time last week at the Epigenetics 2025 meeting from the @gensocuk.bsky.social in sunny Belfast! Great discussions and insights on my PhD project! Also very honored to have been awarded the best poster prize!
Reposted by Thomas Balan
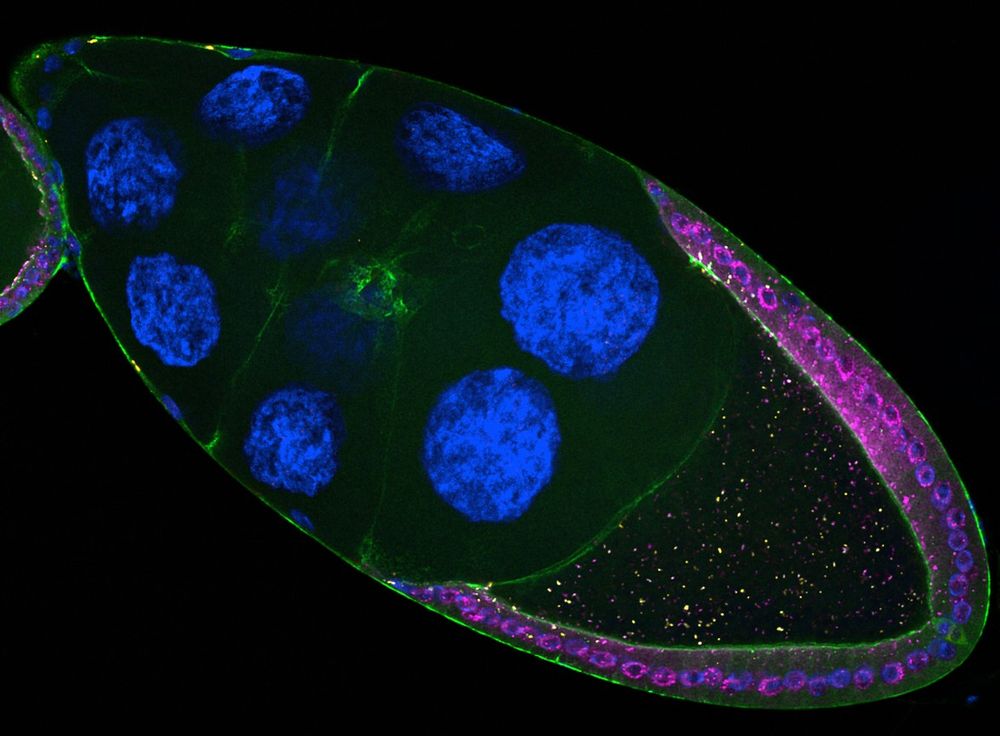
1/ Transposable elements are often called "jumping genes" because they mobilize within genomes. 🧬
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
March 17, 2025 at 11:56 AM
1/ Transposable elements are often called "jumping genes" because they mobilize within genomes. 🧬
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
🚨Cool new study from the lab neighbours! They look into two PAX proteins that orchestrate neuronal diversity!
Dual PAX3/7 transcriptional activities spatially encode spinal cell fates through distinct gene networks https://www.biorxiv.org/content/10.1101/2025.03.11.642668v1
March 18, 2025 at 2:53 PM
🚨Cool new study from the lab neighbours! They look into two PAX proteins that orchestrate neuronal diversity!
Reposted by Thomas Balan
Our paper on Alternative epigenetic states of Arabidopsis Transposable Elements is finally out in Genome Biology !
rdcu.be/d6Yly
rdcu.be/d6Yly
Alternative silencing states of transposable elements in Arabidopsis associated with H3K27me3
rdcu.be
January 22, 2025 at 12:17 PM
Our paper on Alternative epigenetic states of Arabidopsis Transposable Elements is finally out in Genome Biology !
rdcu.be/d6Yly
rdcu.be/d6Yly
Reposted by Thomas Balan

January 13, 2025 at 1:50 PM
Reposted by Thomas Balan
Work led by graduate student @trinitycookis.bsky.social and coauthors Alexandria Lydecker, @paulsauer.bsky.social, and @kasinath-lab.bsky.social is out today where we looked at the inhibition of PRC2 by histone PTMs associated with regions of active transcription.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
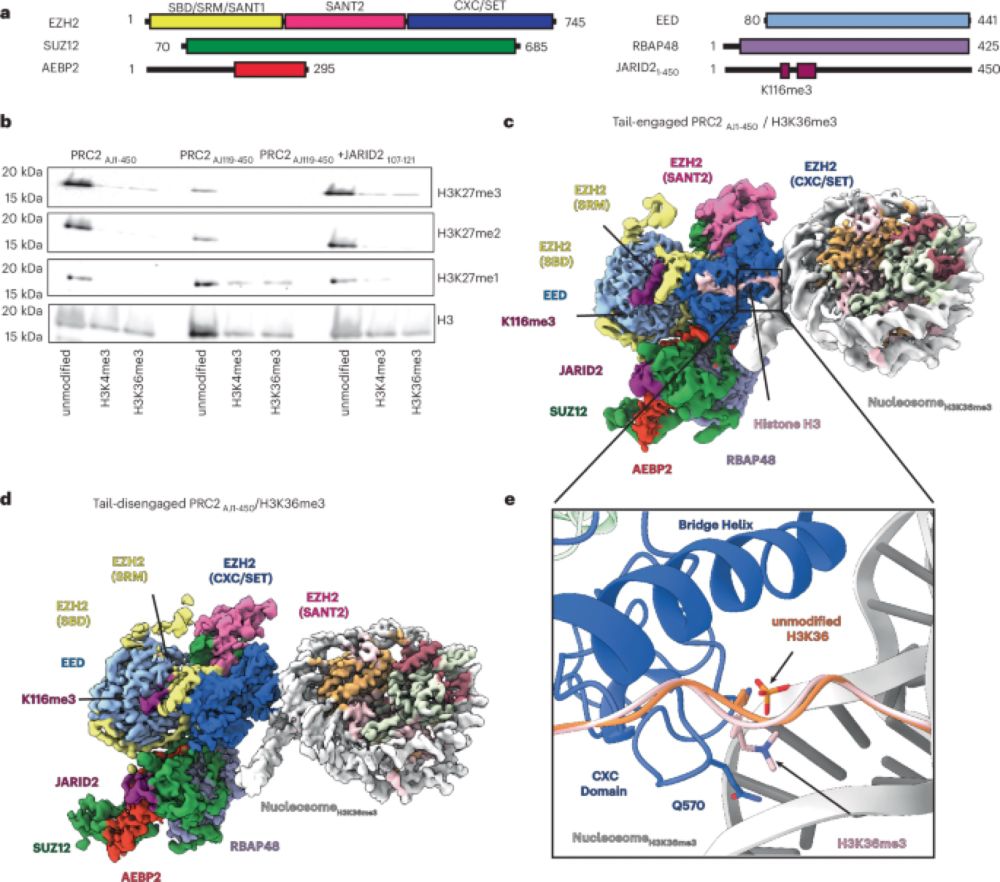
Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications - Nature Structural & Molecular Biology
Structures reveal that histone H3K36me3 and H3K4me3 modifications reduce Polycomb repressive complex 2 (PRC2) function through the inhibition of histone tail engagement and antagonistic binding to the...
www.nature.com
January 8, 2025 at 1:36 AM
Work led by graduate student @trinitycookis.bsky.social and coauthors Alexandria Lydecker, @paulsauer.bsky.social, and @kasinath-lab.bsky.social is out today where we looked at the inhibition of PRC2 by histone PTMs associated with regions of active transcription.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Thomas Balan
A brief overview of my latest project in the Duharcourt lab, highlighted by @cnrs.bsky.social and a sneak peek in the head of an artist (aka my sister) inspired by Paramecium.

Tri sélectif des petits ARN : un enjeu pour préserver l’intégrité du génome
Dans un article publié dans Nucleic Acids Research, des scientifiques ont identifié un nouveau rôle pour la protéine
www.insb.cnrs.fr
December 18, 2024 at 10:19 AM
A brief overview of my latest project in the Duharcourt lab, highlighted by @cnrs.bsky.social and a sneak peek in the head of an artist (aka my sister) inspired by Paramecium.
Reposted by Thomas Balan
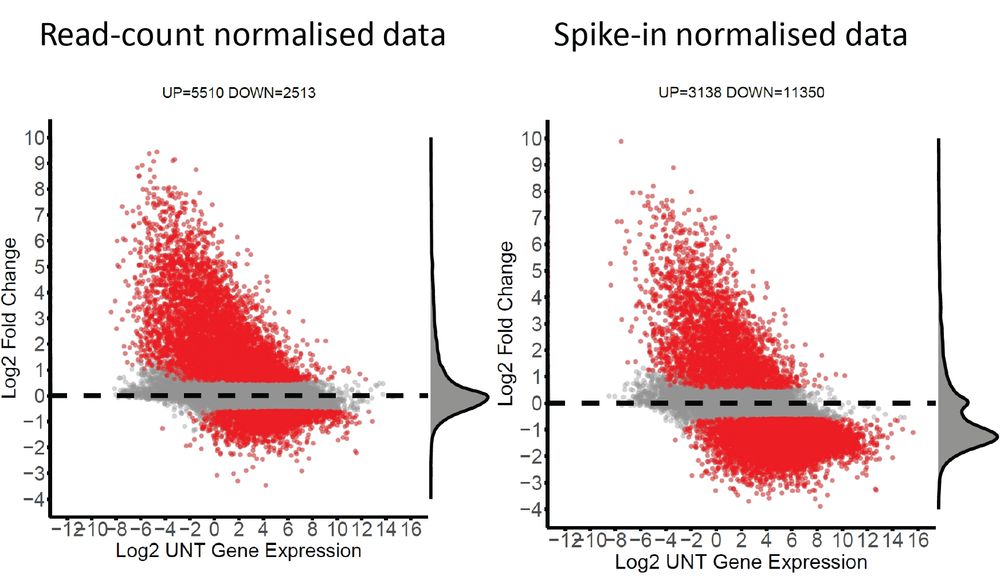
We have had multiple discussions about why people have not previously observed the very dramatic reduction in transcription upon PNUTS depletion. So I looked at our own TT-seq data and analysed it without including the spike-in. The numbers speak for themselves! Spike-in calibration is important!
December 12, 2024 at 10:50 AM
We have had multiple discussions about why people have not previously observed the very dramatic reduction in transcription upon PNUTS depletion. So I looked at our own TT-seq data and analysed it without including the spike-in. The numbers speak for themselves! Spike-in calibration is important!
Very cool new paper from the lab, delving into sRNAs selective degradation enabling TE control! Congrats @oliviacharmant.bsky.social 🎉🎉
For my first post here, I am happy to share that our latest paper is out at #NAR. Thanks for the smooth and constructive reviewing process and the highlight #RNAbiology #TEsky #smallRNAs (1/8)
doi.org/10.1093/nar/...
doi.org/10.1093/nar/...
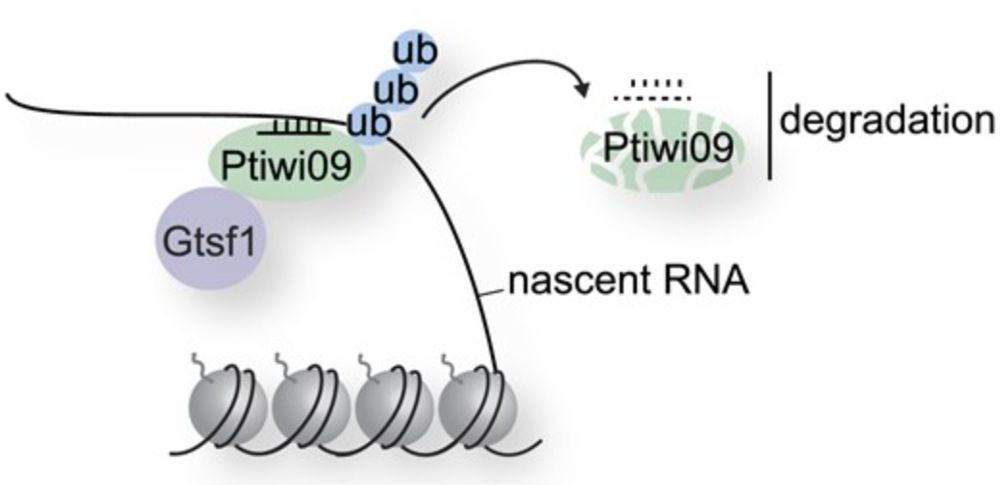
The PIWI-interacting protein Gtsf1 controls the selective degradation of small RNAs in Paramecium
Abstract. Ciliates undergo developmentally programmed genome elimination, in which small RNAs direct the removal of transposable elements (TEs) during the
doi.org
November 25, 2024 at 10:06 AM
Very cool new paper from the lab, delving into sRNAs selective degradation enabling TE control! Congrats @oliviacharmant.bsky.social 🎉🎉
Reposted by Thomas Balan
Please see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator JP Armache! Also big thanks to Mark Foundation for Cancer Research for the support! rdcu.be/dZ5HZ
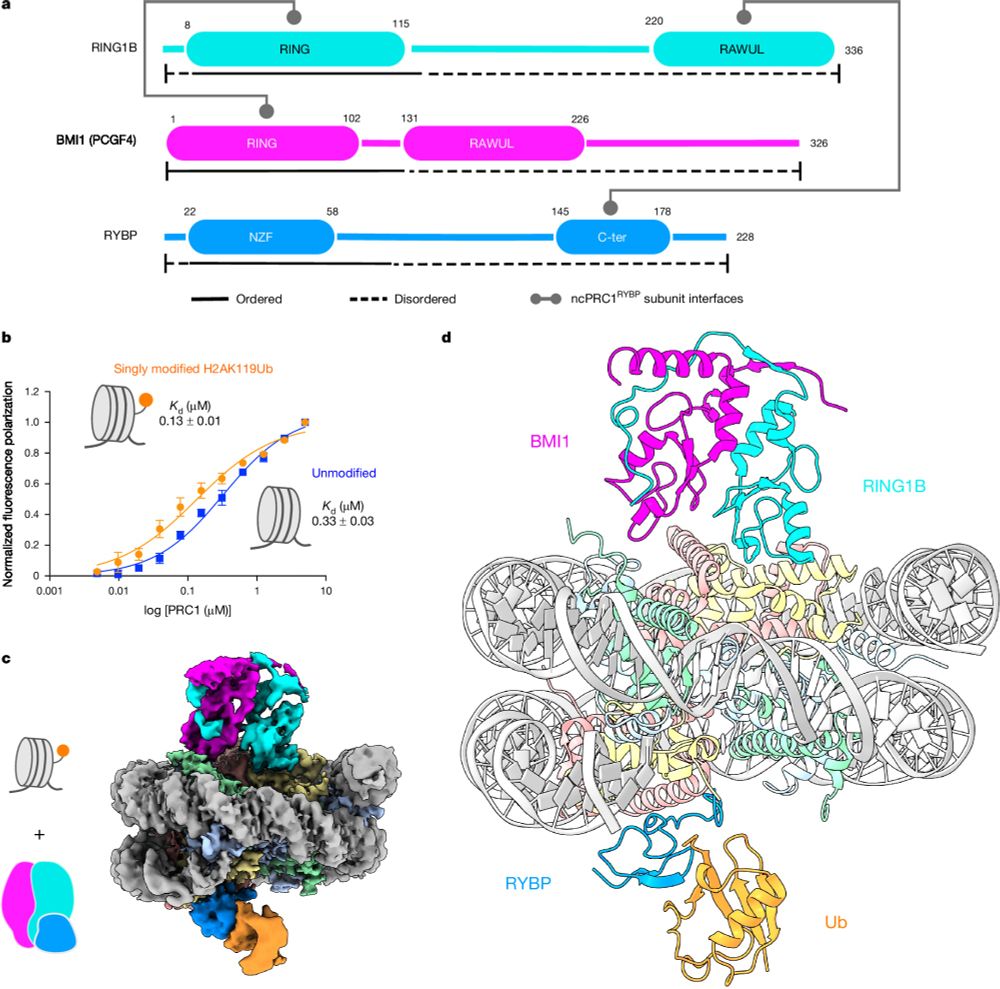
Read–write mechanisms of H2A ubiquitination by Polycomb repressive complex 1
Nature - Cryo-electron microscopy and biochemical studies elucidate the read–write mechanisms of non-canonical PRC1-containing RYBP in histone H2A lysine 119 monoubiquitination and their...
rdcu.be
November 13, 2024 at 6:23 PM
Please see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator JP Armache! Also big thanks to Mark Foundation for Cancer Research for the support! rdcu.be/dZ5HZ

