🔬JB Lalanne on developmental enhancers (and starting a lab in 2025?)
🔬 @rberrens.bsky.social on transposable elements in development
📋Register here: us06web.zoom.us/webinar/regi...

🔬JB Lalanne on developmental enhancers (and starting a lab in 2025?)
🔬 @rberrens.bsky.social on transposable elements in development
📋Register here: us06web.zoom.us/webinar/regi...

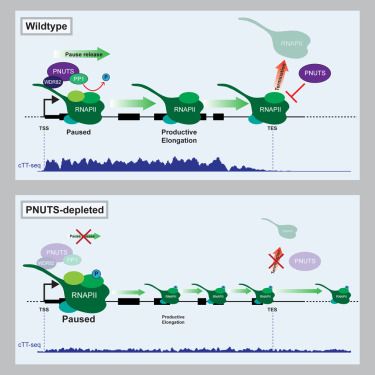
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...

The piece is available open access in Nature Communications:
www.nature.com/articles/s41...

The piece is available open access in Nature Communications:
www.nature.com/articles/s41...
@natsmb.nature.com. A wonderful team effort across the centromere community, across @jansenlab.bsky.social @naltemose.bsky.social @dfachinetti.bsky.social and Giunta labs. Happy reading! 1/4
www.nature.com/articles/s41...

@natsmb.nature.com. A wonderful team effort across the centromere community, across @jansenlab.bsky.social @naltemose.bsky.social @dfachinetti.bsky.social and Giunta labs. Happy reading! 1/4
www.nature.com/articles/s41...
@kashyapchhatbar.bsky.social @saragiuliani.bsky.social @drphcb-uoe.bsky.social @edinburghbiology.bsky.social
@kashyapchhatbar.bsky.social @saragiuliani.bsky.social @drphcb-uoe.bsky.social @edinburghbiology.bsky.social
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.


doi.org/10.1371/jour...
Let's break down what we found! 👇

doi.org/10.1371/jour...
Let's break down what we found! 👇
Hint: Promoter-proximal regions are the key!
Check our latest preprint, led by @mmasoura.bsky.social, to find out!
www.biorxiv.org/content/10.1...

Hint: Promoter-proximal regions are the key!
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
Thanks to @jcellsci.bsky.social for publishing.
journals.biologists.com/jcs/article/...

Thanks to @jcellsci.bsky.social for publishing.
journals.biologists.com/jcs/article/...
Please repost!
jobs.jic.ac.uk/Details.asp?...
Please repost!
jobs.jic.ac.uk/Details.asp?...
www.jic.ac.uk/vacancies/po...
Closing date - 9 November 2025
Salary - £37,500 - £45,350
Contract - 36 months, full-time

www.jic.ac.uk/vacancies/po...
Closing date - 9 November 2025
Salary - £37,500 - £45,350
Contract - 36 months, full-time

www.jic.ac.uk/vacancies/po...
Closing date - 9 November 2025
Salary - £37,500 - £45,350
Contract - 36 months, full-time


www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

register here for the entire series: us06web.zoom.us/webinar/regi...

register here for the entire series: us06web.zoom.us/webinar/regi...


