
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
Slight change to my office window view from Tokyo Tower🗼 to the Tour Eiffel. 🇫🇷

Slight change to my office window view from Tokyo Tower🗼 to the Tour Eiffel. 🇫🇷
Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...

Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...
We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...

We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...
Reference proteomes will remain in UniProtKB, while others will move to UniParc.
Read more about these changes:
www.ebi.ac.uk/about/news/u...
🧬 🖥️
Uniprot is a collaboration between EMBL-EBI, @sib.swiss & the Protein Information Resource.

Reference proteomes will remain in UniProtKB, while others will move to UniParc.
Read more about these changes:
www.ebi.ac.uk/about/news/u...
🧬 🖥️
Uniprot is a collaboration between EMBL-EBI, @sib.swiss & the Protein Information Resource.

Feb 15 - 18, 2026
Brisbane, Australia
Register here: biosig.lab.uq.edu.au/strphy26/reg...
(in-person only)

Feb 15 - 18, 2026
Brisbane, Australia
Register here: biosig.lab.uq.edu.au/strphy26/reg...
(in-person only)
@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz

@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


🌐 evedesign.bio
🌐 evedesign.bio
📄 academic.oup.com/bioinformati...
📄 academic.oup.com/bioinformati...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
This saves some clicking around. Neat!

This saves some clicking around. Neat!
🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social

🚀 search.foldseek.com
foldseek databases Alphafold/UniProt50 afdb50 tmp
foldseek easy-search query.cif afdb50 res.m8 tmp --cluster-search 0/1 (Reps only / all 241M)
Thanks @ebi.embl.org and DeepMind for working with us to make it available to everyone.
Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!

Το συνέδριο #RECOMB2026 θα πραγματοποιηθεί στη Θεσσαλονίκη, στις 26-29 Μαΐου 2026. Οι δορυφορικές εκδηλώσεις θα διεξαχθούν στις 24-25 Μαΐου 2026. Σημειώστε την ημερομηνία!
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
📄 www.nature.com/articles/s41...
💿 mmseqs.com

📄 www.nature.com/articles/s41...
💿 mmseqs.com
www.nature.com/articles/s41...
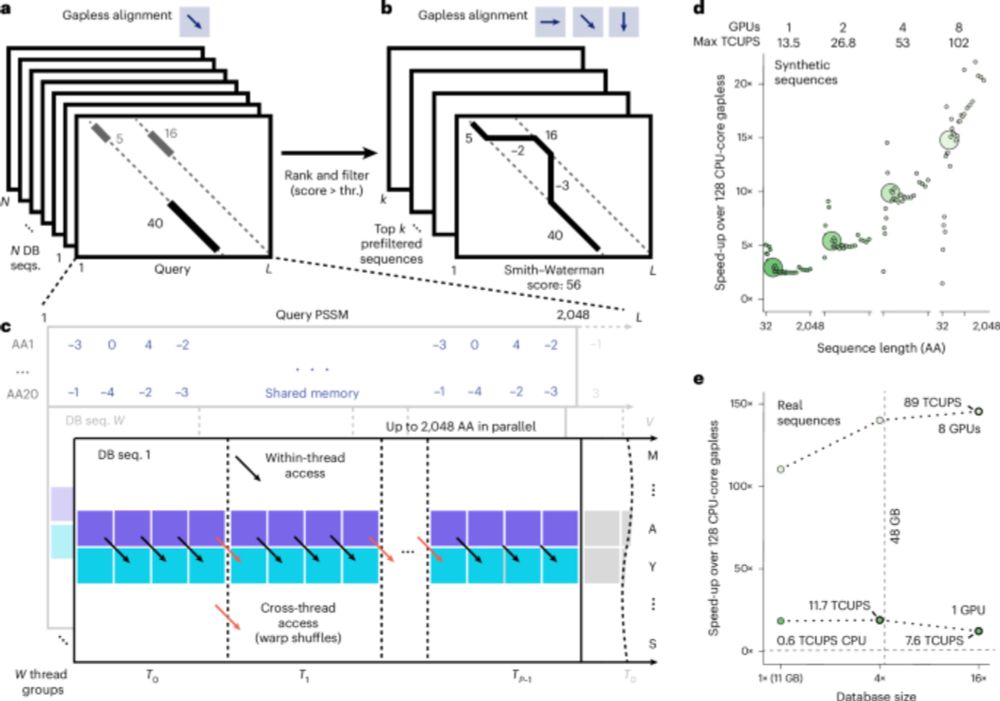
www.nature.com/articles/s41...

