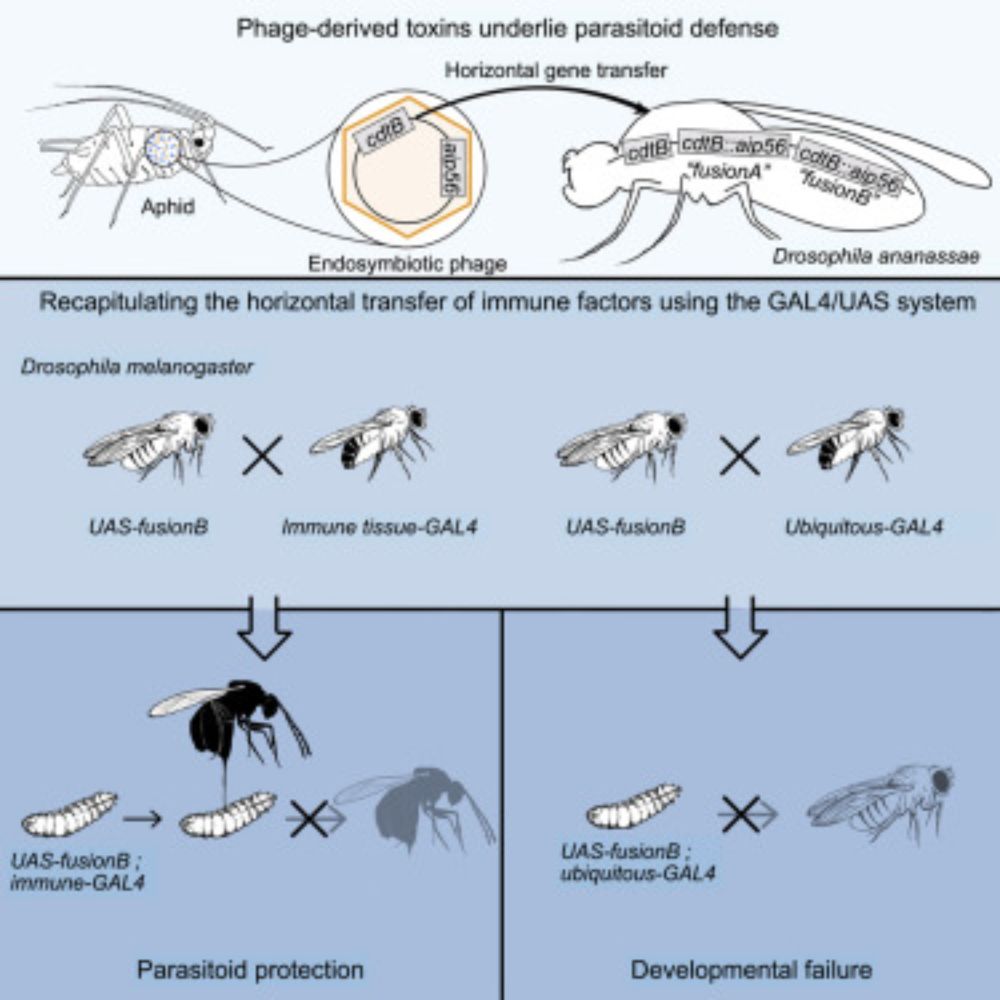
Functional evolution of virus-host interactions. Impact on cross-species transmissions. 🦇 🦍 🦠 🧬 🌳
Genomic and functional #adaptations in #GBP5 highlight specificities of #bat antiviral innate #immunity 🦇🦠
www.biorxiv.org/content/10.1...
✍️ Alexandre Legrand et @lucievirevolte.bsky.social
📕 Nature Ecology and Evolution
buff.ly/Su0EdiU

✍️ Alexandre Legrand et @lucievirevolte.bsky.social
📕 Nature Ecology and Evolution
buff.ly/Su0EdiU
Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...

Evo. characterization #antiviral #SAMD9/9L across #kingdoms🚶♀️🦍🦠🧫🖥️: ancient #convergence + #adaptations @natecoevo.nature.com
Led by amazing Alexandre Legrand +major contributions by Rémi Demeure & Amandine Chantharath @ciri-lyon.bsky.social 1/n
www.nature.com/articles/s41...
sites.google.com/berkeley.edu...


sites.google.com/berkeley.edu...
Find out which early-career researchers will receive funding this year, what they will be investigating, where they will be based... plus lots of other #ERCStG facts & figures for 2025!
➡️ buff.ly/IsafuFh
#FrontierResearch 🇪🇺#EUfunded #HorizonEurope
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
Link: www.nature.com/articles/s41...
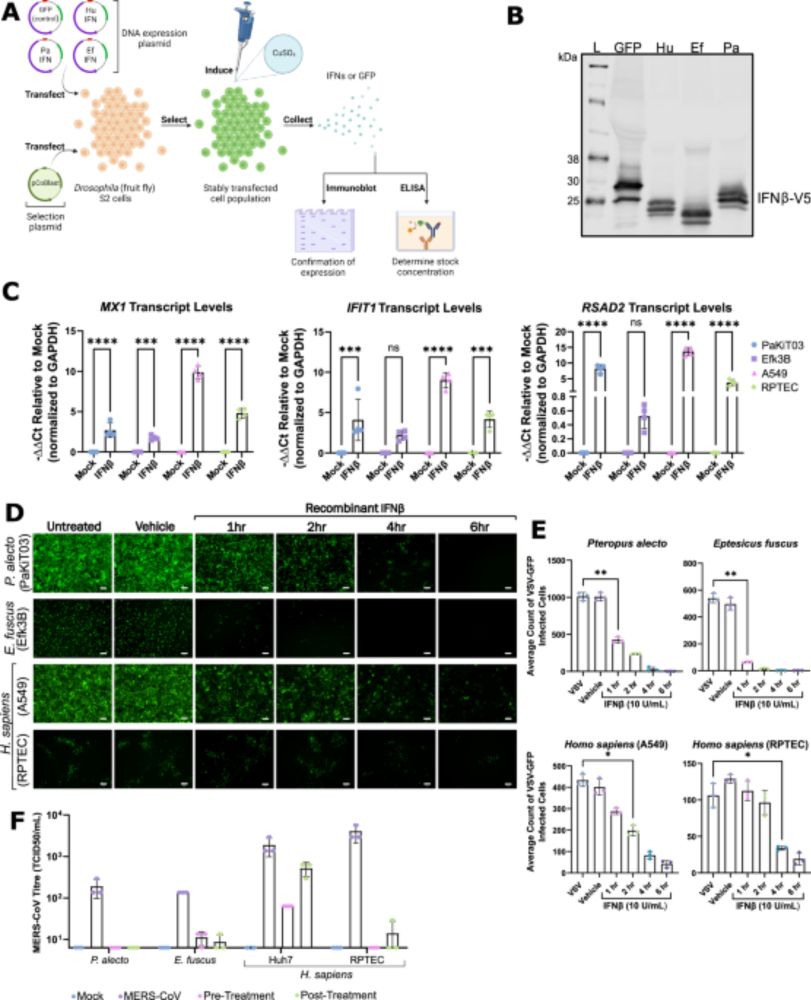
Link: www.nature.com/articles/s41...
For science in France and in the world.
In solidarity to our American colleagues.

For science in France and in the world.
In solidarity to our American colleagues.
*Lyon*
16h00 – Rassemblement place des Terreaux.
*Nancy*
10h00 – Débats. Amphithéâtre de la présidence Léopold, 34 cours Léopold, Nancy.
12-30-13h30 – ronde des obstiné·e·s.
Place Stanislas, Nancy.
*Lyon*
16h00 – Rassemblement place des Terreaux.
*Nancy*
10h00 – Débats. Amphithéâtre de la présidence Léopold, 34 cours Léopold, Nancy.
12-30-13h30 – ronde des obstiné·e·s.
Place Stanislas, Nancy.
Apply now to join us at the IRIM in Montpellier, and lead cutting-edge research in pathogen virulence, host-pathogen interactions or drug resistance, and shape the future of infection biology! 🦠🧫
Please RT (and follow us!)
🗓️ Deadline: Feb. 28, 2025

Apply now to join us at the IRIM in Montpellier, and lead cutting-edge research in pathogen virulence, host-pathogen interactions or drug resistance, and shape the future of infection biology! 🦠🧫
Please RT (and follow us!)
🗓️ Deadline: Feb. 28, 2025
@cnrsbiologie.bsky.social @lucievirevolte.bsky.social @ensdelyon.bsky.social @inserm.fr
www.youtube.com/watch?v=jE8b...

@cnrsbiologie.bsky.social @lucievirevolte.bsky.social @ensdelyon.bsky.social @inserm.fr
www.youtube.com/watch?v=jE8b...
The sub-cellular localization of important innate antiviral factor GBP5 has evolved multiple times during bat evolution, with consequences for activity.
www.biorxiv.org/content/10.1...
The sub-cellular localization of important innate antiviral factor GBP5 has evolved multiple times during bat evolution, with consequences for activity.
www.biorxiv.org/content/10.1...
The sub-cellular localization of important innate antiviral factor GBP5 has evolved multiple times during bat evolution, with consequences for activity.
www.biorxiv.org/content/10.1...
Genomic and functional #adaptations in #GBP5 highlight specificities of #bat antiviral innate #immunity 🦇🦠
www.biorxiv.org/content/10.1...
Genomic and functional #adaptations in #GBP5 highlight specificities of #bat antiviral innate #immunity 🦇🦠
www.biorxiv.org/content/10.1...
doi.org/10.1093/gbe/...
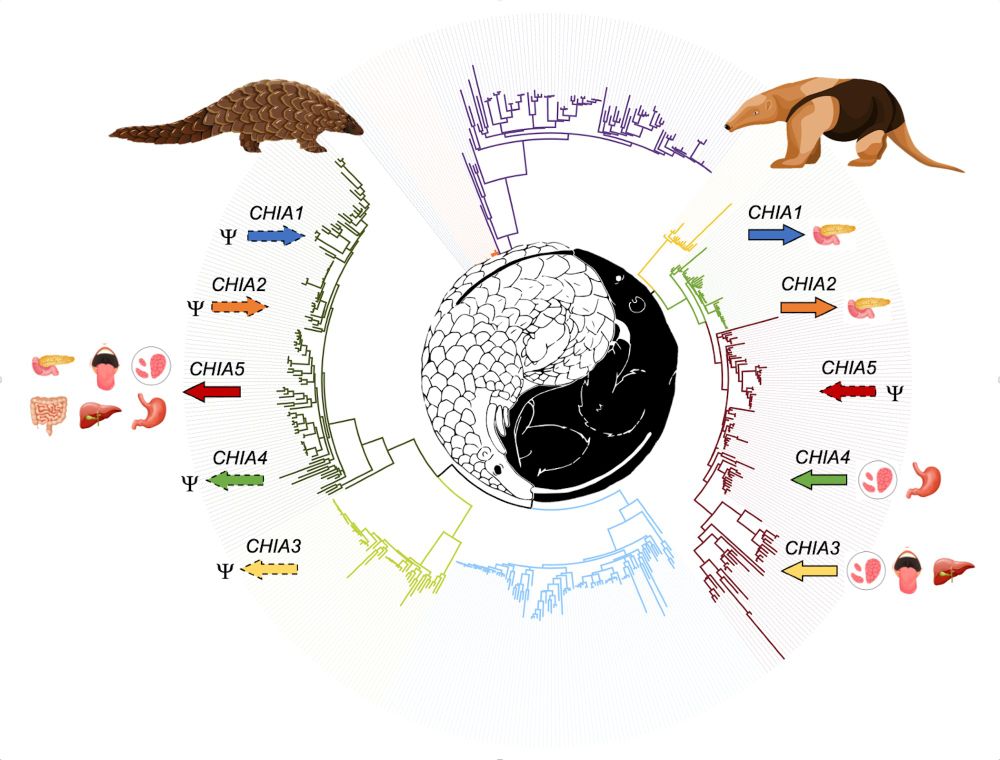
doi.org/10.1093/gbe/...
Your mentorship is incredible - Thank you


Your mentorship is incredible - Thank you
#EvoCure #comparativeImmunology #virology
Please share
@inserm.fr @ciri-lyon.bsky.social @cnrsbiologie.bsky.social
ciri.ens-lyon.fr/teams/lp2l/j...
#EvoCure #comparativeImmunology #virology
Please share
@inserm.fr @ciri-lyon.bsky.social @cnrsbiologie.bsky.social
ciri.ens-lyon.fr/teams/lp2l/j...
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...
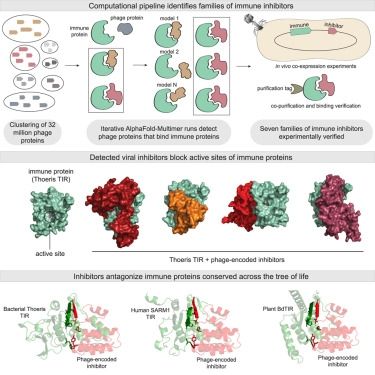
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...
www.science.org/doi/10.1126/...
In this project co-led with @ostermanilya.bsky.social at @soreklab.bsky.social , we show that a bacterial immune system employs a TIR protein and a caspase-like protease, two typical immune components seen in eukaryotes. 🧵👇
www.science.org/doi/10.1126/...
In this project co-led with @ostermanilya.bsky.social at @soreklab.bsky.social , we show that a bacterial immune system employs a TIR protein and a caspase-like protease, two typical immune components seen in eukaryotes. 🧵👇
doi.org/10.1093/nar/...
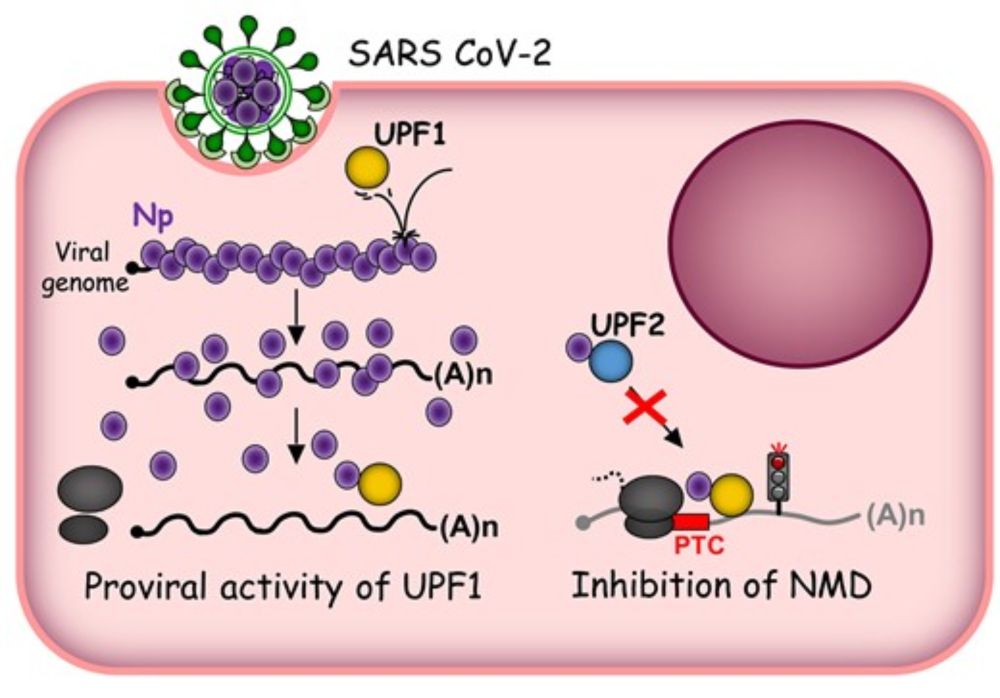
doi.org/10.1093/nar/...
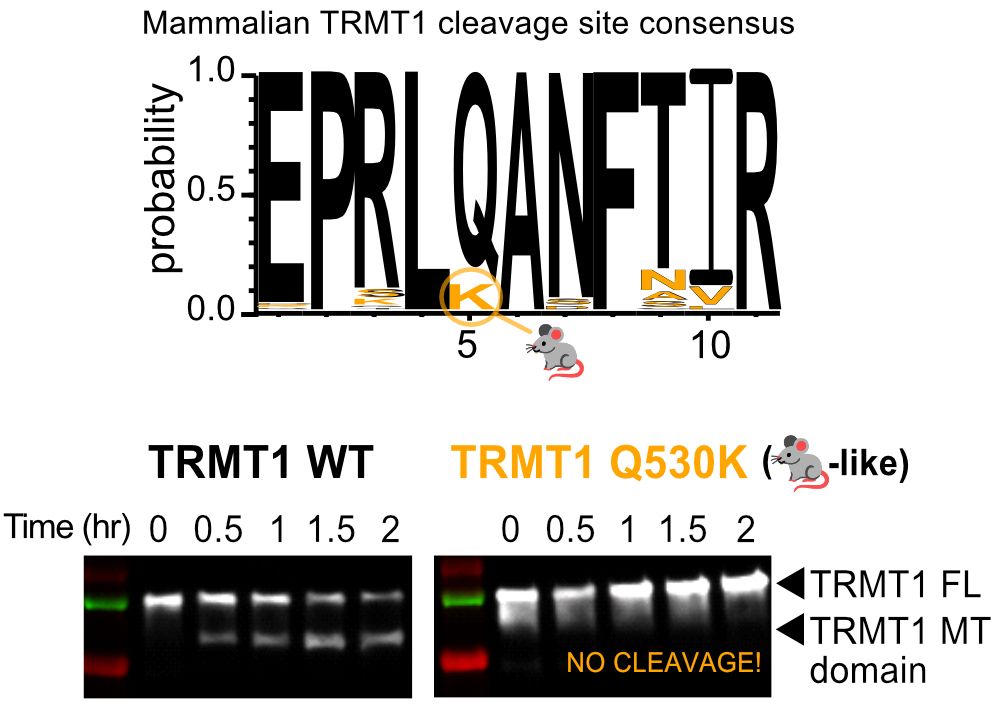
💡TRMT1 cleavage & inactivation may be another mechanism by which SARS-CoV-2 manipulates host tRNA modifications and translation!
🦠We think this could impact viral pathogenesis or phenotypes!
💊Our structure might help understand Mpro substrate recognition or different pockets to drug!
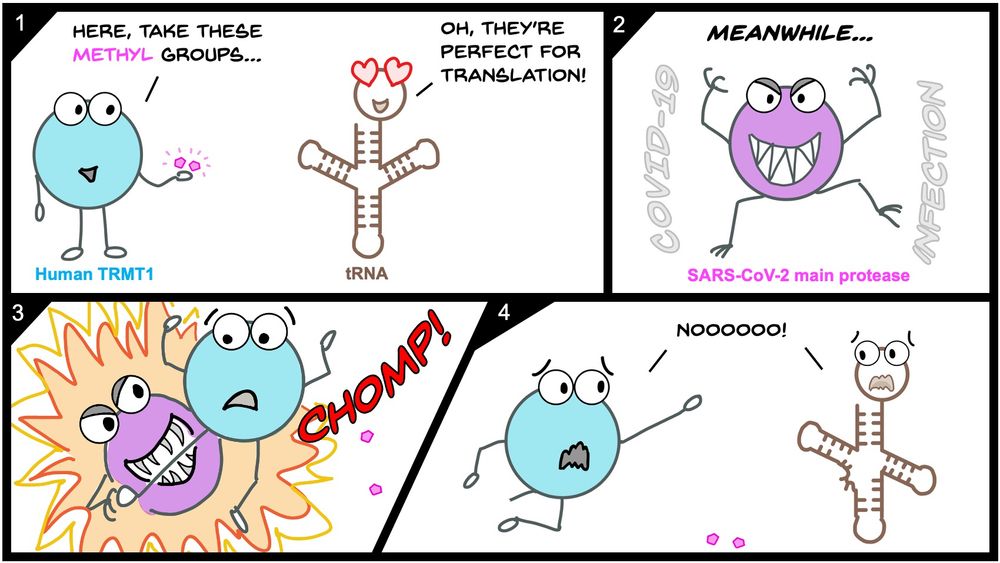
💡TRMT1 cleavage & inactivation may be another mechanism by which SARS-CoV-2 manipulates host tRNA modifications and translation!
🦠We think this could impact viral pathogenesis or phenotypes!
💊Our structure might help understand Mpro substrate recognition or different pockets to drug!
elifesciences.org/articles/91168
A short bluetorial... 🧵 1/9

elifesciences.org/articles/91168
A short bluetorial... 🧵 1/9
We're thrilled to unwrap Viro3D - a comprehensive database of virus protein structures: >85,000 predicted structures from 4,400 human & animal viruses! 🦠
viro3d.cvr.gla.ac.uk
We're thrilled to unwrap Viro3D - a comprehensive database of virus protein structures: >85,000 predicted structures from 4,400 human & animal viruses! 🦠
viro3d.cvr.gla.ac.uk